Metabolomic and Transcriptomic Analysis Reveal the Role of Metabolites and Genes in Modulating Flower Color of Paphiopedilum micranthum
Abstract
:1. Introduction
2. Results
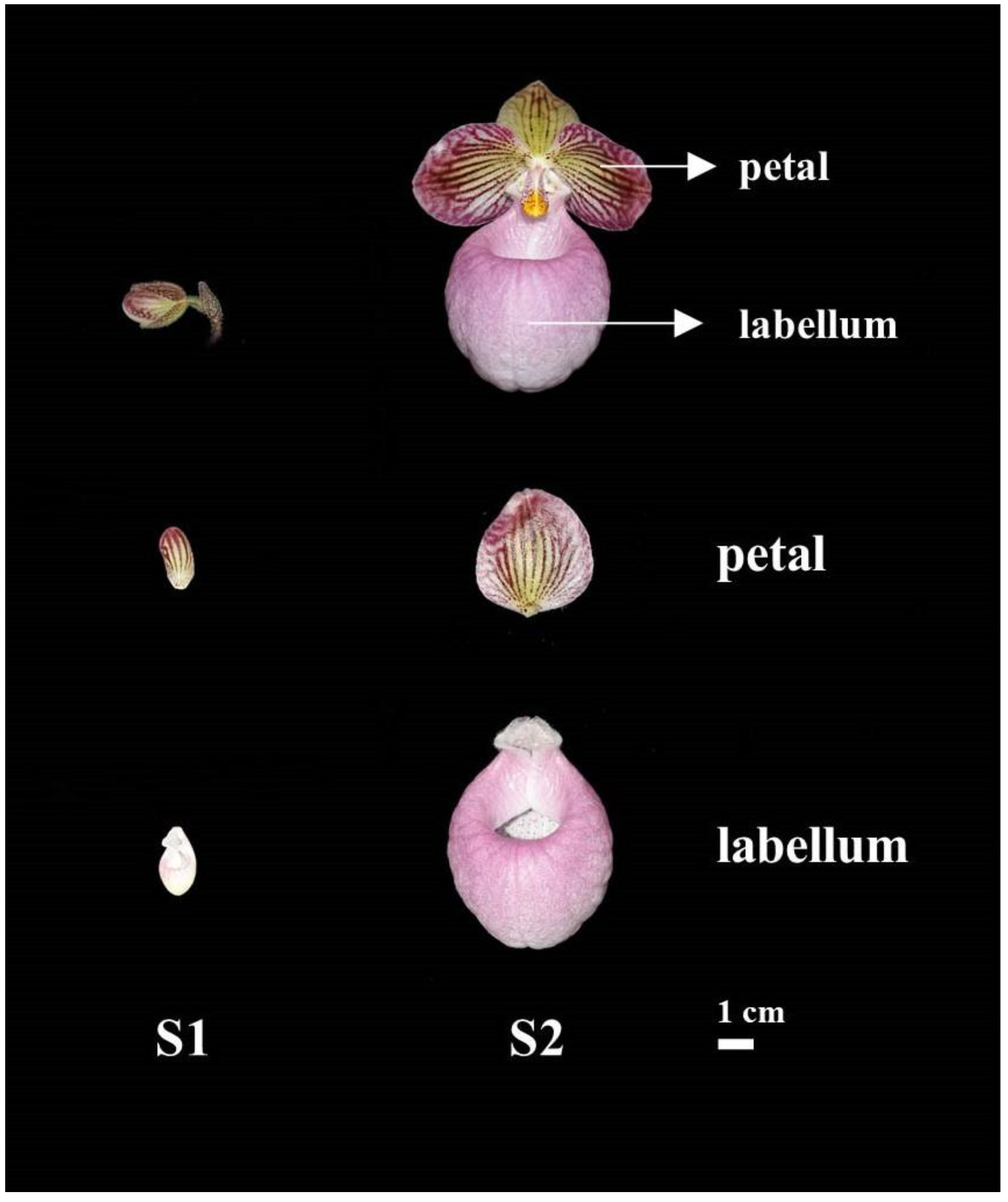
2.1. Morphology and Phenotypic Modifications of P. micranthum during Floral Development
2.2. Identification and Quantification of Pigment Components
2.3. Differentially Expressed Genes Related to the Pigment Biosynthesis
2.4. DEGs Involved in the ABP and CBP
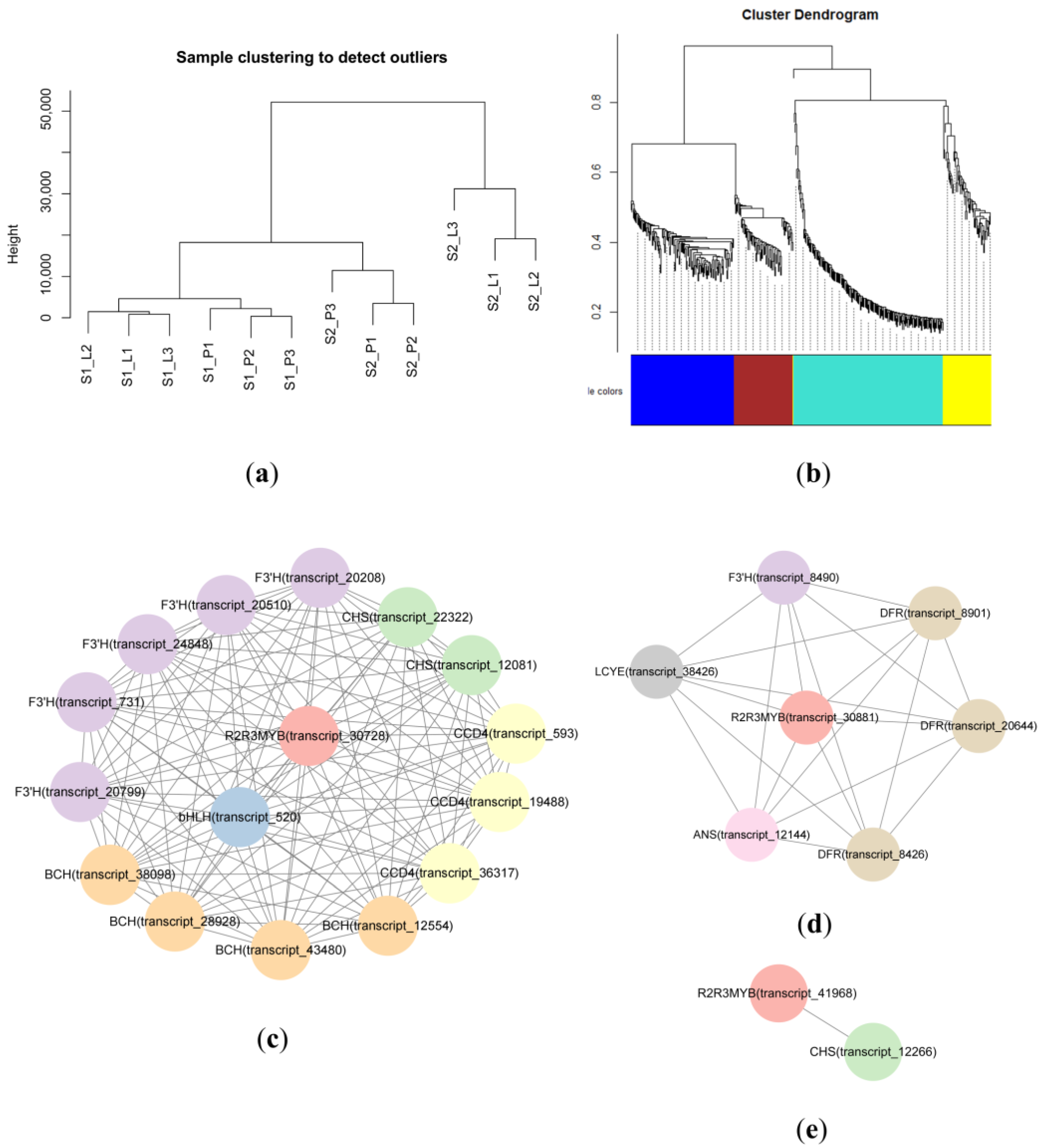
2.5. Weighted Gene Coexpression Network Analysis (WGCNA)
3. Discussion
3.1. Different Concentration of Accumulated Pigments Instigate Phenotypic Modifications in P. micranthum
3.2. Different Accumulation of Pigments Related to the Expression of the Structural Genes
3.3. R2R3-MYBs and bHLH Regulate the Structural Genes of ABP and CBP
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. Pigment Separation
4.3. Targeted Metabolome Profiling and Analysis
4.4. RNA Extraction, Library Preparation, Sequencing, and Annotation
4.5. DEGs, Functional Enrichment Analysis, and WGCNA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willmer, P. Pollination and Floral Ecology; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Jersáková, J.; Johnson, S.D.; Jürgens, A. Food deception by plants: From generalized systems to specialized floral mimicry. In Plant-Environment Interactions; Balu¿ka, F., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 223–246. [Google Scholar]
- Gumbert, A.; Kunze, J. Colour similarity to rewarding model plants affects pollination in a food deceptive orchid, Orchis boryi. Biol. J. Linn. Soc. 2001, 72, 419–433. [Google Scholar] [CrossRef]
- Ma, X.; Shi, J.; Bänziger, H.; Sun, Y.; Guo, Y.; Liu, Z.; Luo, Y. The functional significance of complex floral colour pattern in a food-deceptive orchid. Funct. Ecol. 2016, 30, 721–732. [Google Scholar] [CrossRef]
- Galizia, C.G.; Kunze, J.; Gumbert, A.; Borg-Karlson, A.K.; Sachse, S.; Markl, C.; Menzel, R. Relationship of visual and olfactory signal parameters in a food-deceptive flower mimicry system. Behav. Ecol. 2005, 16, 159–168. [Google Scholar] [CrossRef]
- Kunze, J.; Gumbert, A. The combined effect of color and odor on flower choice behavior of bumble bees in flower mimicry systems. Behav. Ecol. 2001, 12, 447–456. [Google Scholar] [CrossRef]
- Luo, H.; Liang, Y.; Xiao, H.; Liu, N.; Chen, Y.; Wang, W.; Tang, J.; Xiong, D.; Yang, B.; Ren, Z. Deceptive pollination of Calanthe by skippers that commonly act as nectar thieves. Entomol. Sci. 2020, 23, 3–9. [Google Scholar] [CrossRef]
- Policha, T.; Davis, A.; Barnadas, M.; Dentinger, B.T.; Raguso, R.A.; Roy, B.A. Disentangling visual and olfactory signals in mushroom-mimicking Dracula orchids using realistic three-dimensional printed flowers. New Phytol. 2016, 210, 1058–1071. [Google Scholar] [CrossRef]
- Niggebrugge, C.; Leboulle, G.; Menzel, R.; Komischke, B.; de Ibarra, N.H. Fast learning but coarse discrimination of colours in restrained honeybees. J. Exp. 2009, 212, 1344–1350. [Google Scholar] [CrossRef]
- Salzmann, C.C.; Nardella, A.M.; Cozzolino, S.; Schiestl, F.P. Variability in floral scent in rewarding and deceptive orchids: The signature of pollinator-imposed selection? Ann. Bot. 2007, 100, 757–765. [Google Scholar] [CrossRef]
- Peter, C.I.; Johnson, S.D. Mimics and magnets: The importance of color and ecological facilitation in floral deception. Ecology 2008, 89, 1583–1595. [Google Scholar] [CrossRef]
- Jersáková, J.; Jürgens, A.; Šmilauer, P.; Johnson, S.D. The evolution of floral mimicry: Identifying traits that visually attract pollinators. Funct. Ecol. 2012, 26, 1381–1389. [Google Scholar] [CrossRef]
- Fenster, C.B.; Armbruster, W.S.; Wilson, P.; Dudash, M.R.; Thomson, J.D. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 375–403. [Google Scholar] [CrossRef]
- Fantinato, E.; Del Vecchio, S.; Baltieri, M.; Fabris, B.; Buffa, G. Are food-deceptive orchid species really functionally specialized for pollinators? Ecol. Res. 2017, 32, 951–959. [Google Scholar] [CrossRef]
- Phillips, R.D.; Peakall, R.; van der Niet, T.; Johnson, S.D. Niche perspectives on plant–pollinator interactions. Trends Plant Sci. 2020, 25, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Van der Kooi, C.J.; Elzenga, J.T.M.; Staal, M.; Stavenga, D.G. How to colour a flower: On the optical principles of flower coloration. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160429. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Schlüter, P.M.; Schiestl, F.P. Molecular mechanisms of floral mimicry in orchids. Trends Plant Sci. 2008, 13, 228–235. [Google Scholar] [CrossRef]
- Zhang, R.; Lu, Y. Molecular mechanisms and natural selection of flower color variation. Bot. Res. 2016, 5, 186–209. [Google Scholar]
- Wessinger, C.A.; Rausher, M.D. Lessons from flower colour evolution on targets of selection. J. Exp. Bot. 2012, 63, 5741–5749. [Google Scholar] [CrossRef]
- Sun, T.; Li, L. Toward the “golden” era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2019, 290, 110331. [Google Scholar] [CrossRef]
- Farzad, M.; Griesbach, R.; Hammond, J.; Weiss, M.R.; Elmendorf, H.G. Differential expression of three key anthocyanin biosynthetic genes in a color-changing flower, Viola cornuta cv. Yesterday, Today and Tomorrow. Plant Sci. 2003, 1651, 1333–1342. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A. Diversity of carotenoid composition in flower petals. Jpn. Agric. Res. Q. JARQ 2011, 45, 163–171. [Google Scholar] [CrossRef]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, Z.; Wang, Y.; Wang, C.; Zhu, B.; Liu, H. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell 2019, 31, 2751–2767. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.W. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Renner, S.S. Rewardless flowers in the angiosperms and the role of insect cognition in their evolution. In Plant-Pollinator Interactions: From Specialization to Generalization; Waser, N.M., Ollerton, J., Eds.; University of Chicago Press: Chicago, MA, USA, 2006; pp. 123–144. [Google Scholar]
- Jersáková, J.; Johnson, S.D.; Kindlmann, P. Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev. 2006, 81, 219–235. [Google Scholar] [CrossRef]
- Cozzolino, S.; Widmer, A. Orchid diversity: An evolutionary consequence of deception? Trends Ecol. Evol. 2005, 20, 487–494. [Google Scholar] [CrossRef]
- Shrestha, M.; Dyer, A.G.; Dorin, A.; Ren, Z.X.; Burd, M. Rewardlessness in orchids: How frequent and how rewardless? Plant Biol. 2020, 22, 555–561. [Google Scholar] [CrossRef]
- Scaccabarozzi, D.; Cozzolino, S.; Guzzetti, L.; Galimberti, A.; Milne, L.; Dixon, K.W.; Phillips, R.D. Masquerading as pea plants: Behavioural and morphological evidence for mimicry of multiple models in an Australian orchid. Ann. Bot. 2018, 122, 1061–1073. [Google Scholar] [CrossRef]
- Scaccabarozzi, D.; Lunau, K.; Guzzetti, L.; Cozzolino, S.; Dyer, A.G.; Tommasi, N.; Gagliano, M. Mimicking orchids lure bees from afar with exaggerated ultraviolet signals. Ecol. Evol. 2023, 13, e9759. [Google Scholar] [CrossRef]
- Liang, C.Y.; Rengasamy, K.P.; Huang, L.M.; Hsu, C.C.; Jeng, M.F.; Chen, W.H.; Chen, H.H. Assessment of violet-blue color formation in Phalaenopsis orchids. BMC Plant Biol. 2020, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.C.; Perkins, J.; Peakall, R. Anthocyanin and Flavonol Glycoside Metabolic Pathways Underpin Floral Color Mimicry and Contrast in a Sexually Deceptive Orchid. Front. Plant Sci. 2022, 13, 860997. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, K.; Takamura, T.; Fukai, S. Effects of high temperature on flower colour and anthocyanin content in pink flower genotypes of greenhouse chrysanthemum (Chrysanthemum morifolium Ramat). J. Hortic. Sci. Biotechnol. 2006, 81, 728–734. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Dong, B.; Fu, J.; Hu, S.; Zhao, H. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans. Front. Plant Sci. 2018, 9, 1499. [Google Scholar] [CrossRef]
- Chiou, C.Y.; Pan, H.A.; Chuang, Y.N.; Yeh, K.W. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 2010, 232, 937–948. [Google Scholar] [CrossRef]
- Hsiao, Y.Y.; Huang, T.H.; Fu, C.H.; Huang, S.C.; Chen, Y.J.; Huang, Y.M.; Chen, H.H. Transcriptomic analysis of floral organs from Phalaenopsis orchid by using oligonucleotide microarray. Gene 2013, 518, 91–100. [Google Scholar] [CrossRef]
- Han, Y.; Wang, X.; Chen, W.; Dong, M.; Yuan, W.; Liu, X.; Shang, F. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet. Genomes 2014, 10, 329–338. [Google Scholar] [CrossRef]
- Yamamizo, C.; Kishimoto, S.; Ohmiya, A. Carotenoid composition and carotenogenic gene expression during Ipomoea petal development. J. Exp. Bot. 2010, 61, 709–719. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.H. Integrative Metabolomic and Transcriptome Analysis Reveal the Differential Mechanisms of Spot Color in the Lips of Dendrobium chrysotoxum. J. Plant Biol. 2022, 1–13. [Google Scholar] [CrossRef]
- Liu, H.; Kishimoto, S.; Yamamizo, C.; Fukuta, N.; Ohmiya, A. Carotenoid accumulations and carotenogenic gene expressions in the petals of Eustoma grandiflorum. Plant Breed. 2013, 132, 417–422. [Google Scholar] [CrossRef]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. FPB 2012, 39, 619–638. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.W.; Duan, Y.W.; Men, S.; Wang, L. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Li, B.J.; Zheng, B.Q.; Wang, J.Y.; Tsai, W.C.; Lu, H.C.; Zou, L.H. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, X.; Hu, C.; Xie, C.; Lu, A.; Luo, Y.; Peng, T.; Huang, W. Metabolomic and Transcriptomic Analysis Reveal the Role of Metabolites and Genes in Modulating Flower Color of Paphiopedilum micranthum. Plants 2023, 12, 2058. https://doi.org/10.3390/plants12102058
Deng X, Hu C, Xie C, Lu A, Luo Y, Peng T, Huang W. Metabolomic and Transcriptomic Analysis Reveal the Role of Metabolites and Genes in Modulating Flower Color of Paphiopedilum micranthum. Plants. 2023; 12(10):2058. https://doi.org/10.3390/plants12102058
Chicago/Turabian StyleDeng, Xinyan, Chao Hu, Chengzhi Xie, Aixian Lu, Yibo Luo, Tao Peng, and Weichang Huang. 2023. "Metabolomic and Transcriptomic Analysis Reveal the Role of Metabolites and Genes in Modulating Flower Color of Paphiopedilum micranthum" Plants 12, no. 10: 2058. https://doi.org/10.3390/plants12102058
APA StyleDeng, X., Hu, C., Xie, C., Lu, A., Luo, Y., Peng, T., & Huang, W. (2023). Metabolomic and Transcriptomic Analysis Reveal the Role of Metabolites and Genes in Modulating Flower Color of Paphiopedilum micranthum. Plants, 12(10), 2058. https://doi.org/10.3390/plants12102058




