Biofilm-Forming Ability of Phytopathogenic Bacteria: A Review of its Involvement in Plant Stress
Abstract
1. Introduction
2. Abiotic Stress in Plants and Its Relation to Pathogenesis
2.1. Salt Stress
2.2. Drought Stress
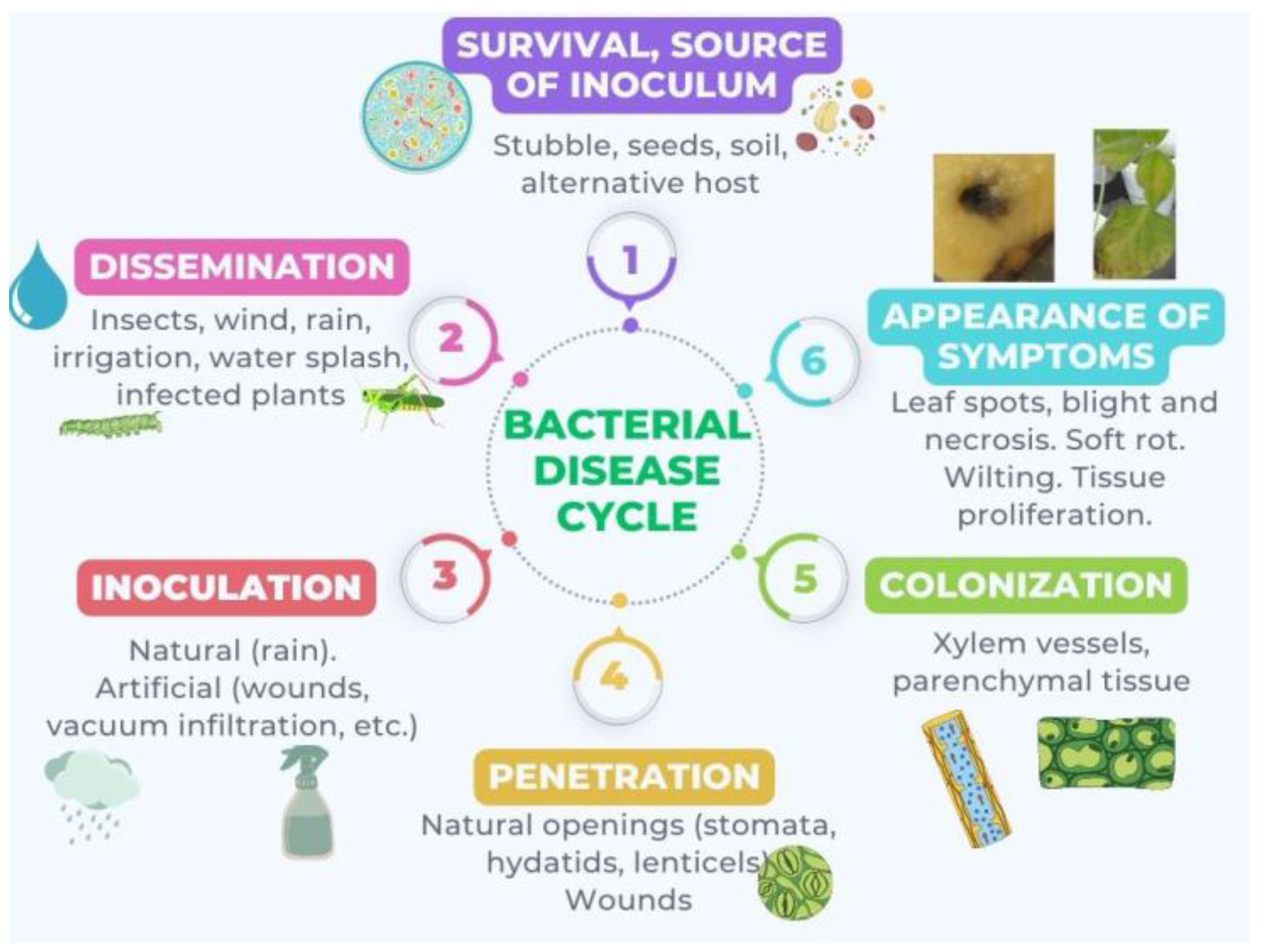
3. Plant Resistance to Biotic Stress and Survival/Infection Strategies by Phytopathogenic Bacteria
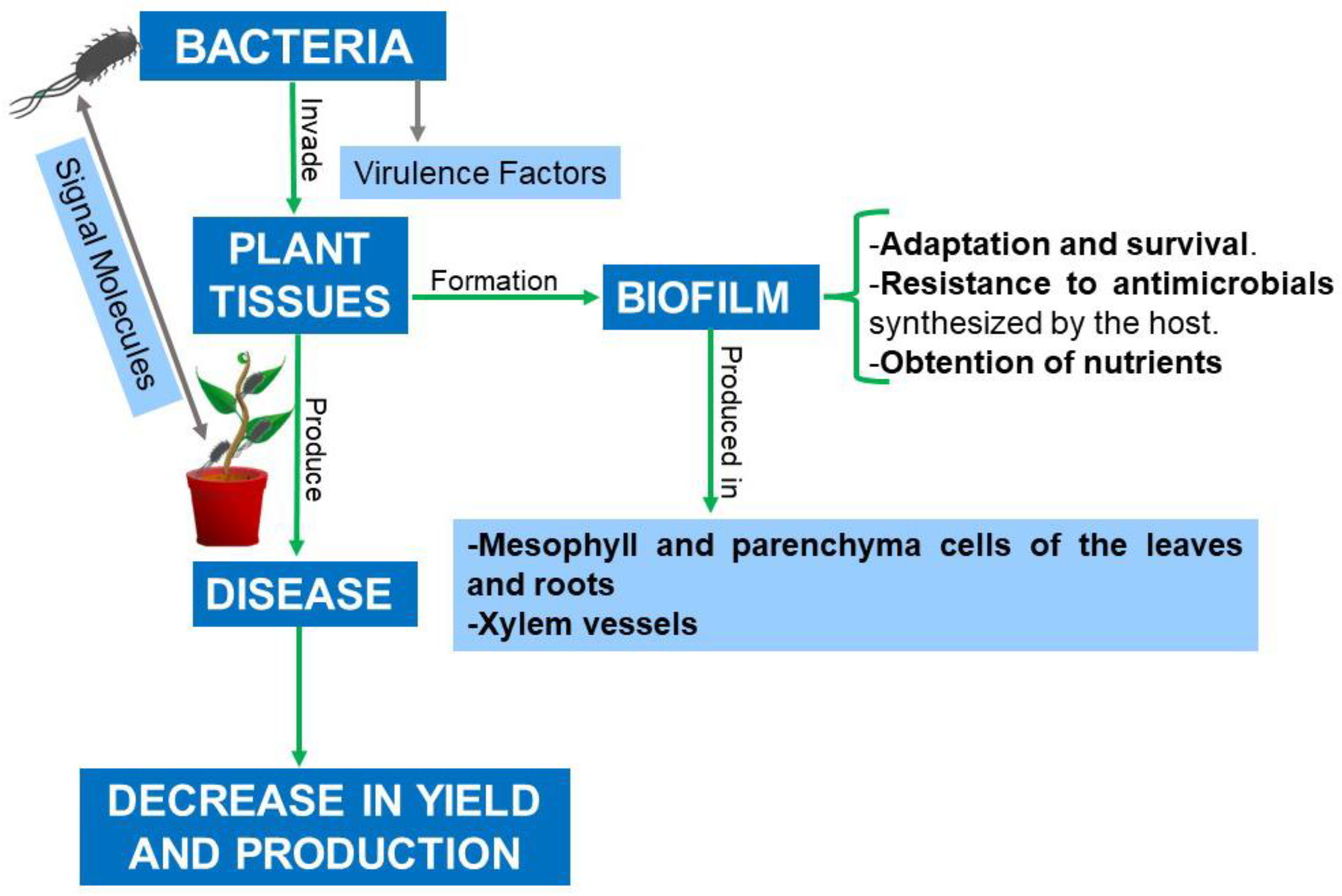
4. Biofilm: Composition, Functions, and Stages of Formation
- Adhesion: the microorganisms engage in weak interactions (acid-base, hydrophobic, Van der Waals, and electrostatic forces) to reversibly adhere to a surface.
- Colonization: irreversible bonds come about through hydrophilic/hydrophobic interactions; the bacteria use flagella, pili, and collagen-binding adhesive proteins.
- Development: EPS are secreted and there is a continuous proliferation and accumulation of cells.
- Maturation: the three-dimensional structure settles into its stable form featuring circulation and signaling channels.
5. Social Behavior of the Bacterial Population in the Biofilm Matrix and Its Relationship with Pathogenicity
6. Biofilm in Plants
7. Biofilm Formed by Phytopathogenic Bacteria
7.1. Phytopathogenic Bacteria that Colonize Xylem Vessels
7.2. Phytopathogenic Bacteria that Colonize Root Tissues
7.3. Phytopathogenic Bacteria that Colonize Parenchymal Tissues
8. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lambers, H.; Chapin, F.S.; Pons, T.L. Plant Physiological Ecology; Springer-Verlag: New York, NY, USA, 1998; p. 540. [Google Scholar]
- Kumari, S.; Vaishnav, A.; Jain, S.; Varma, A.; Choudhary, D.K. Induced drought tolerance through wild and mutant bacterial strain Pseudomonas simiae in mung bean (Vigna radiata L.). World J. Microbiol. Biotechnol. 2016, 32, 4. [Google Scholar] [CrossRef]
- Schulze, E.D.; Beck, E.; Buchmann, N.; Clemens, S.; Müller-Hohenstein, K.; Scherer-Lorenzen, M. Water deficiency (Drought). In Plant Ecology; Schulze, E.-D., Beck, E., Buchmann, N., Clemens, S., Müller-Hohenstein, K., Scherer-Lorenzen, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 165–202. [Google Scholar] [CrossRef]
- Chiappero, J.; Cappellari, L.R.; Palermo, T.B.; Giordano, W.; Khan, N.; Banchio, E. Plant growth promoting rhizobacteria affect the antioxidant status in medicinal and aromatic plants grown under osmotic-stress. Ind. Crops Prod. 2021, 167, 113541. [Google Scholar] [CrossRef]
- Martins, P.M.; Merfa, M.V.; Takita, M.A.; De Souza, A.A. Persistence in phytopathogenic bacteria: Do we know enough? Front. Microbiol. 2018, 9, 1099. [Google Scholar] [CrossRef]
- Khan, M.; Khan, A.U.; Hasan, M.A.; Yadav, K.K.; Pinto, M.M.C.; Malik, N.; Yadav, V.K.; Khan, A.H.; Islam, S.; Sharma, G.K. Agro-nanotechnology as an emerging field: A novel sustainable approach for improving plant growth by reducing biotic stress. Appl. Sci. 2021, 11, 2282. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.F.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Recent advances in bacterial amelioration of plant drought and salt stress. Biology 2022, 11, 437. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Stavi, I.; Thevs, N.; Priori, S. Soil salinity and sodicity in drylands: A review of causes, effects, monitoring, and restoration measures. Front. Environ. Sci. 2021, 9, 712831. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Liu, X.M.; Zhang, H. The effects of bacterial volatile emissions on plant abiotic stress tolerance. Front. Plant Sci. 2015, 6, 774. [Google Scholar] [CrossRef]
- Mithofer, A.; Schulze, B.; Boland, W. Biotic and heavy metal stress response in plants: Evidence for common signals. FEBS Lett. 2004, 566, 1–5. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Physiol. 2014, 164, 1636–1648. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Reg. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Triky-Dotan, S.; Yermiyahu, U.; Katan, J.; Gamliel, A. Development of crown and root rot disease of tomato under irrigation with salinewater. Phytopathology 2005, 95, 1438–1444. [Google Scholar] [CrossRef]
- DiLeo, M.V.; Pye, M.F.; Roubtsova, T.V.; Duniway, J.M.; MacDonald, J.D.; Rizzo, D.M.; Bostock, R.M. Abscisic acid in salt stress predis-position to Phytophthora root and crown rot in tomato and chrysanthemum. Phytopathology 2010, 100, 871–879. [Google Scholar] [CrossRef]
- Wiese, J.; Kranz, T.; Schubert, S. Induction of pathogen resistance in barley by abiotic stress. Plant Biol. 2004, 6, 529–536. [Google Scholar] [CrossRef]
- Lipiec, J.; Doussan, C.; Nosalewicz, A.; Kondracka, K. Effect of drought and heat stresses on plant growth and yield: A review. Int. Agrophys. 2013, 27, 463–477. [Google Scholar] [CrossRef]
- Vandana, U.K.; Singha, B.; Gulzar, A.B.M.; Mazumder, P.B. Molecular Mechanisms in Plant Growth Promoting Bacteria (PGPR) to Resist Environmental Stress in Plants. In Molecular Aspects of Plant Beneficial Microbes in Agriculture; Sharma, V., Salwan, R., Al-Ani, L.K.T., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 221–233. [Google Scholar] [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833834. [Google Scholar] [CrossRef]
- Chandra, P.; Wunnava, A.; Verma, P.; Chandra, A.; Sharma, R.K. Strategies to mitigate the adverse effect of drought stress on crop plants-influences of soil bacteria: A review. Pedosphere 2021, 31, 496–509. [Google Scholar] [CrossRef]
- Huang, B.; Chen, Y.-E.; Zhao, Y.-Q.; Ding, C.-B.; Liao, J.-Q.; Hu, C.; Zhou, L.-J.; Zhang, Z.-W.; Yuan, S.; Yuan, M. Exogenous melatonin alleviates oxidative damages and protects photosystem ii in maize seedlings under drought stress. Front. Plant Sci. 2019, 10, 677. [Google Scholar] [CrossRef]
- Zafra, A.; Rodríguez-García, M.I.; Alch’e, J.d.D. Cellular localization of ROS and NO in olive reproductive tissues during flower development. BMC Plant Biol. 2010, 10, 36. [Google Scholar] [CrossRef]
- Ngumbi, E.; Kloepper, J. Bacterial-mediated drought tolerance: Current and future prospects. Appl. Soil Ecol. 2016, 105, 109–125. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Achuo, E.A.; Prinsen, E.; Höfte, M. Influence of drought, salt stress and abscisic acid on the resistance of tomato to Botrytis cinerea and Oidium neolycopersici. Plant Pathol. 2006, 55, 178–186. [Google Scholar] [CrossRef]
- Prasch, C.M.; Sonnewald, U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef]
- Diourte, M.; Starr, J.L.; Jeger, M.J.; Stack, J.P.; Rosenow, D.T. Charcoal rot (Macrophomina phaseolina) resistance and the effects of water stress on disease development in sorghum. Plant Pathol. 1995, 44, 196–202. [Google Scholar] [CrossRef]
- Mayek-Pérez, N.; GarcÍa-Espinosa, R.; López-Castañeda, C.; Acosta-Gallegos, J.A.; Simpson, J. Water relations, histopathology and growth of common bean (Phaseolus vulgaris L.) during pathogenesis of Macrophomina phaseolina under drought stress. Physiol. Mol. Plant Pathol. 2002, 60, 185–195. [Google Scholar] [CrossRef]
- McElrone, A.J.; Sherald, J.L.; Forseth, I.N. Effects of water stress on symptomatology and growth of Parthenocissus quinquefolia infected by Xylella fastidiosa. Plant Dis. 2001, 85, 1160–1164. [Google Scholar] [CrossRef]
- Suleman, P.; Al-Musallam, A.; Menezes, C.A. The effect of solute potential and water stress on black scorch caused by Chalara paradoxa and Chalara radicicola on date palms. Plant Dis. 2001, 85, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, N.; Raghav, M.; Dubey, S.; Bedi, N. Bacterial exopolysaccharides: Insight into their role in plant abiotic stress tolerance. J. Microbiol. Biotechnol. 2021, 31, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, E.; Salinas, M.; Manzano-Agugliaro, F. Worldwide research on plant defense against biotic stresses as an improvement for sustainable agriculture. Sustainability 2018, 10, 391. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Azizullah, L.D.; Manzoor, I.; Song, F. Plant-microbiome crosstalk: Dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int. J. Mol. Sci. 2021, 22, 6852. [Google Scholar] [CrossRef]
- Plant Pathology, 5th ed.; Academic Press: Cambridge, MA, USA, 2005; (Edición en castellano, Ed. LIMUSA).
- Van der Wolf, J.; De Boer, S. Phytopathogenic Bacteria. In Principles of Plant-Microbe Interactions; Lugtenberg, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Zehra, A.; Raytekar, N.A.; Meena, M.; Swapnil, P. Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: A review. Curr. Res. Microb. Sci. 2021, 2, 100054. [Google Scholar] [CrossRef]
- Moustafa-Farag, M.; Almoneafy, A.; Mahmoud, A.; Elkelish, A.; Arnao, M.B.; Li, L.; Ai, S. Melatonin and its protective role against biotic stress impacts on plants. Biomolecules 2020, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Mosqueda, M.C.; Fadiji, A.E.; Babalola, O.O.; Santoyo, G. Bacterial elicitors of the plant immune system: An overview and the way forward. Plant Stress 2023, 7, 100138. [Google Scholar] [CrossRef]
- Li, P.; Lu, Y.J.; Chen, H.; Day, B. The lifecycle of the plant immune system. CRC Crit. Rev. Plant Sci. 2020, 39, 72–100. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Microbiol. Spectr. 2016, 4, 213–239. [Google Scholar] [CrossRef]
- Morgan, J.L.W.; Acheson, J.F.; Zimmer, J. Structure of a type-1 secretion system ABC transporter. Structure 2017, 25, 522–529. [Google Scholar] [CrossRef]
- Alav, I.; Kobylka, J.; Kuth, M.S.; Klaas, M.; Pos, M.P.; Blair, J.M.A.; Bavro, V.N. Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 2021, 121, 5479–5596. [Google Scholar] [CrossRef]
- Yamazaki, A.; Hirata, H.; Tsuyumu, S. HrpG regulates type II secretory proteins in Xanthomonas axonopodis pv. citri. J. Gen. Plant Pathol. 2008, 74, 138–150. [Google Scholar] [CrossRef]
- Korotkov, K.V.; Sandkvist, M. Architecture, function, and substrates of the type II secretion system. EcoSal Plus 2019, 8, 2. [Google Scholar] [CrossRef]
- Wagner, S.; Grin, I.; Malmsheimer, S.; Singh, N.E.C.; Westerhausen, S. Bacterial type III secretion systems: A complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol. Lett. 2018, 365, fny201. [Google Scholar] [CrossRef]
- Yuan, X.; Yu, M.; Yang, C.-H. Innovation and application of the type III secretion system inhibitors in plant pathogenic bacteria. Microorganisms 2020, 8, 1956. [Google Scholar] [CrossRef]
- Büttner, D.; He, S.Y. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009, 150, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Puhar, A.; Sansonetti, P.J. Type III secretion system. Curr. Biol. 2014, 24, R784–R791. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, M.R.; Anderson, J.C. Regulation of the Pseudomonas syringae Type III secretion system by host environment signals. Microorganisms 2021, 9, 1227. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef]
- Nahar, K.; Matsumoto, I.; Taguchi, F.; Inagaki, Y.; Yamamoto, M.; Toyoda, K.; Shiraishi, T.; Ichinose, Y.; Mukaihara, T. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 2014, 15, 297–303. [Google Scholar] [CrossRef]
- Lonjon, F.; Turner, M.; Henry, C.; Rengel, D.; Lohou, D.; van de Kerkhove, Q.; Cazalé, A.C.; Peeters, N.; Genin, S.; Vailleau, F. Comparative secretome analysis of Ralstonia solanacearum Type 3 secretion-associated mutants reveals a fine control of effector delivery, essential for bacterial pathogenicity. MCP 2016, 15, 598–613. [Google Scholar] [CrossRef]
- Cascales, E.; Christie, P.J. Definition of a bacterial Type IV secretion pathway for a DNA substrate. Science 2004, 304, 1170–1173. [Google Scholar] [CrossRef]
- Low, H.; Gubellini, F.; Rivera-Calzada, A.; Braun, N.; Connery, S.; Dujeancourt, A.; Lu, F.; Redzej, A.; Fronzes, R.; Orlova, E.V.; et al. Structure of a type IV secretion system. Nature 2014, 508, 550–553. [Google Scholar] [CrossRef]
- Alvarez-Martinez, C.E.; Christie, P.J. Biological diversity of prokaryotic type IV secretion systems. MMBR 2009, 73, 775–808. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Martinez, C.E.; Sgro, G.G.; Araujo, G.G.; Paiva, M.R.N.; Matsuyama, B.Y.; Guzzo, C.R.; Andrade, M.O.; Farah, C.S. Secrete or perish: The role of secretion systems in Xanthomonas biology. Comput. Struct. Biotechnol. J. 2021, 19, 279–302. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R.; Hor, L.; Pilapitiya, A.; Luirink, J.; Paxman, J.J.; Heras, B. Phylogenetic classification and functional review of autotransporters. Front. Immunol. 2022, 13, 921272. [Google Scholar] [CrossRef]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 2018, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed]
- Borrero de Acuña, J.M.; Bernal, P. Interacciones de holobiontes vegetales mediadas por el sistema de secreción tipo VI y las vesículas de membrana: Herramientas prometedoras para una agricultura más verde. Environ. Microbiol. 2021, 23, 1830–1836. [Google Scholar] [CrossRef] [PubMed]
- Lomovatskaya, L.A.; Romanenko, A.S. Secretion systems of bacterial phytopathogens and mutualists (Review). Appl. Biochem. Microbiol. 2020, 56, 115–129. [Google Scholar] [CrossRef]
- Lauber, F.; Deme, J.C.; Lea, S.M.; Berks, B.C. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 2018, 564, 77–82. [Google Scholar] [CrossRef]
- Bogino, P.; Abod, A.; Nievas, F.; Giordano, W. Water-limiting conditions alter the structure and biofilm-forming ability of bacterial multispecies communities in the alfalfa rhizosphere. PLoS ONE 2013, 8, e79614. [Google Scholar] [CrossRef]
- Liaqat, I.; Liaqat, M.; Tahir, H.M.; Haq, I.; Ali, N.M.; Arshad, M.; Arshad, N. Motility effects biofilm formation in Pseudomonas aeruginosa and Enterobacter cloacae. Pak. J. Pharm. Sci. 2019, 32, 927–932. [Google Scholar]
- Beoletto, V.G.; De Las Mercedes Oliva, M.; Marioli, J.M.; Carezzano, M.E.; Demo, M.S. Antimicrobial natural products against bacterial biofilms. In Antibiotic Resistance: Mechanisms and New Antimicrobial Approaches; Kon, K., Rai, M., Eds.; Elsevier: London, UK, 2016; pp. 290–307. [Google Scholar]
- Srinivasan, R.; Santhakumari, S.; Poonguzhali, P.; Geetha, M.; Dyavaiah, M.; Xiangmin, L. Bacterial biofilm inhibition: A focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front. Microbiol. 2021, 12, 676458. [Google Scholar] [CrossRef]
- Masák, J.; Čejková, A.; Schreiberová, O.; Rezanka, T. Pseudomonas biofilms: Possibilities of their control. FEMS Microbiol. Ecol. 2014, 89, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489. [Google Scholar] [CrossRef]
- Castiblanco, L.F.; Sundin, G.W. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J. Integr. Plant Biol. 2016, 58, 362–372. [Google Scholar] [CrossRef]
- Baltenneck, J.; Reverchon, S.; Hommais, F. Regulación de detección de quórum en bacterias fitopatógenas. Microorganismos 2021, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Preda, V.G.; Săndulescu, O. Communication is the key: Biofilms, quorum sensing, formation and prevention. Discoveries 2019, 7, e100. [Google Scholar] [CrossRef]
- Li, J.; Zhao, X. Effects of quorum sensing on the biofilm formation and viable but non-culturable state. Food Res. Int. 2020, 137, 109742. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Satpathy, S.; Kumar Sen, S.; Pattanaik, S.; Raut, S. Review on bacterial biofilm: An universal cause of contamination. Biocatal. Agric. Biotechnol. 2016, 7, 56–66. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.; Greenberg, E. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Atkinson, S.; Williams, P. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 2009, 6, 959–978. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef]
- Zhou, J.; Cai, Z. Microbial Social Interactions in Biofilm. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Bramhachari, P.V., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 29–46. [Google Scholar] [CrossRef]
- Velmourougane, K.; Prasanna, R.; Saxena, A.K. Agriculturally important microbial biofilms: Present status and future prospects. J. Basic Microbiol. 2017, 57, 548–573. [Google Scholar] [CrossRef]
- Diggle, S.P.; Gardner, A.; West, S.A.; Griffin, A.S. Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philos. Trans, R. Soc. Lond B. Biol. Sci. 2007, 362, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Donati, I.; Fiorentini, L.; Vandelle, E.; Polverari, A.; Venturi, V.; Buriani, G.; Vanneste, J.L.; Spinelli, F. N-Acyl Homoserine lactones and lux solos regulate social behaviour and virulence of Pseudomonas syringae pv. actinidiae. Microb. Ecol. 2020, 79, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Nadell, C.D.; Xavier, J.B.; Foster, K.R. The sociobiology of biofilms. FEMS Microbiology Reviews 2009, 33, 206–224. [Google Scholar] [CrossRef]
- Li, Y.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538. [Google Scholar] [CrossRef]
- Morris, C.E.; Monier, J.M. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 2003, 41, 429–453. [Google Scholar] [CrossRef]
- Patwardhan, S.B.; Pandit, C.; Pandit, S.; Verma, D.; Lahiri, D.; Nag, M.; Ray, R.R.; Jha, P.; Prasad, R. Illuminating the signalomics of microbial biofilm on plant surfaces, Biocatal. Agric. Biotechnol. 2023, 47, 102537. [Google Scholar] [CrossRef]
- Rafique, M.; Hayat, K.; Mukhtar, T.; Khan, A.A.; Afridi, M.S.; Hussain, T.; Sultan, T.; Munis, M.F.H.; Imran, M.; Chaudhary, H.J. Bacterial Biofilm Formation and Its Role Against Agricultural Pathogens. In The Battle against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; Mendez Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2015; pp. 373–382. [Google Scholar]
- Ajijah, N.; Fiodor, A.; Pandey, A.K.; Rana, A.; Pranaw, K. Plant Growth-Promoting Bacteria (PGPB) with biofilm-forming ability: A multifaceted agent for sustainable agriculture. Diversity 2023, 15, 112. [Google Scholar] [CrossRef]
- Rudrappa, T.; Bais, H.P. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008, 56, 1955–1962. [Google Scholar] [CrossRef] [PubMed]
- Mina, I.R.; Jara, N.P.; Criollo, J.E.; Castillo, J.A. The critical role of biofilms in bacterial vascular plant pathogenesis. Plant Pathol. 2019, 68, 1439–1447. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393. [Google Scholar] [CrossRef] [PubMed]
- Tomihama, T.; Nishi, Y.; Arai, K. Biofilm formation and resistance to bactericides of Pseudomonas syringae pv. theae. J. Gen. Plant Pathol. 2006, 73, 193–196. [Google Scholar] [CrossRef]
- Sidorova, D.E.; Skripka, M.I.; Khmel, I.A.; Koksharova, O.A.; Plyuta, V.A. Effects of volatile organic compounds on biofilms and swimming motility of Agrobacterium tumefaciens. Microorganisms 2022, 10, 1512. [Google Scholar] [CrossRef]
- Zecharia, N.; Krasnov, H.; Vanunu, M.; Siri, A.C.; Haberman, A.; Dror, O.; Vakal, L.; Almeida, R.P.P.; Blank, L.; Shtienberg, D.; et al. Xylella fastidiosa Outbreak in Israel: Population genetics, host range, and temporal and spatial distribution analysis. Phytopathology 2022, 11, 2296–2309. [Google Scholar] [CrossRef]
- Dagher, F.; Nickzad, A.; Zheng, J.; Hoffmann, M.; Déziel, E. Characterization of the biocontrol activity of three bacterial isolates against the phytopathogen Erwinia amylovora. Microbiology 2021, 10, e1202. [Google Scholar] [CrossRef]
- Sowndarya, J.; Rubini, D.; Sinsinwar, S.; Senthilkumar, M.; Nithyanand, P.; Vadivel, V. Gallic acid an agricultural byproduct modulates the biofilm matrix exopolysaccharides of the phytopathogen Ralstonia solanacearum. Curr. Microbiol. 2020, 77, 3339–3354. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yuan, Z.-H.; Zhang, H.; Pan, Y.; Wu, Y.; Tian, X.-Q.; Wang, F.-F.; Wang, L.; Qian, W. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog. 2017, 13, e1006304. [Google Scholar] [CrossRef] [PubMed]
- Ndemueda, A.; Pereira, I.; Faustino, M.A.F.; Cunha, Â. Photodynamic inactivation of the phytopathogenic bacterium Xanthomonas citri subsp. citri. Lett. Appl. Microbiol. 2020, 4, 420–427. [Google Scholar] [CrossRef]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The role of quorum sensing molecules in bacterial–plant interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, S.; Moleleki, L.N.; Shyntum, D.Y.; Coutinho, T.A. Quorum Sensing in Gram-Negative Plant Pathogenic Bacteria. In Advances in Plant Pathology; Kimatu, J.N., Ed.; IntechOpen: London, UK, 2018; Volume 10. [Google Scholar] [CrossRef]
- Merighi, M.; Majerczak, D.R.; Coplin, D.L. A novel transcriptional autoregulatory loop enhances expression of the Pantoea stewartii subsp. stewartii Hrp type III secretion system. FEMS Microbiol. Lett. 2005, 243, 479–487. [Google Scholar] [CrossRef]
- Yuan, X.; Hulin, M.T.; Sundin, G.W. Effectors, chaperones, and harpins of the Type III secretion system in the fire blight pathogen Erwinia amylovora: A review. J. Plant Pathol. 2021, 103, 25–39. [Google Scholar] [CrossRef]
- Liu, F.; Hu, M.; Zhang, Z.; Xue, Y.; Chen, S.; Hu, A.; Zhang, L.-h.; Zhou, J. Dickeya manipulates multiple quorum sensing systems to control virulence and collective behaviors. Front Plant Sci. 2022, 13, 68. [Google Scholar] [CrossRef]
- Hossain, A.; Abdallah, Y.; Ali, M.A.; Masum, M.M.I.; Li, B.; Sun, G.; Meng, Y.; Wang, Y.; An, Q. Lemon-fruit-based green synthesis of zinc oxide nanoparticles and titanium dioxide nanoparticles against soft rot bacterial pathogen Dickeya dadantii. Biomolecules 2019, 9, 863. [Google Scholar] [CrossRef]
- Hossain, A.; Hong, X.; Ibrahim, E.; Li, B.; Sun, G.; Meng, Y.; Wang, Y.; An, Q. Green synthesis of silver nanoparticles with culture supernatant of a bacterium Pseudomonas rhodesiae and their antibacterial activity against soft rot pathogen Dickeya dadantii. Molecules 2019, 24, 2303. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, J.; Yang, Q.; Sun, D.; Pu, X.; Shen, H.; Li, Q.; Wang, Z.; Lin, B. Antimicrobial activity of natural plant compound carvacrol against soft rot disease agent Dickeya zeae. Curr. Microbiol. 2021, 78, 3453–3463. [Google Scholar] [CrossRef]
- Hajian-Maleki, H.; Sareh, B.-R.; Mohammad, M. Efficiency of essential oils against Pectobacterium carotovorum subsp. carotovorum causing potato soft rot and their possible application as coatings in storage. Postharvest Biol. Technol. 2019, 156, 110928. [Google Scholar] [CrossRef]
- Li, B.; Huang, J.; Yi, Y.; Liu, S.; Liu, R.; Xiao, Z.; Li, C. Effects of rhapontigenin as a novel quorum-sensing inhibitor on exoenzymes and biofilm formation of Pectobacterium carotovorum subsp. carotovorum and its application in vegetables. Molecules 2022, 27, 8878. [Google Scholar] [CrossRef] [PubMed]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Feitosa-Junior, O.R.; Souza, A.P.S.; Zaini, P.A.; Baccari, C.; Ionescu, M.; Pierry, P.M.; Uceda-Campos, G.; Labroussaa, F.; Almeida, R.P.P.; Lindow, S.E.; et al. The XadA trimeric autotransporter adhesins in Xylella fastidiosa differentially contribute to cell aggregation, biofilm formation, insect transmission and virulence to plants. Mol. Plant-Microbe Interact. 2022, 35, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Roper, M.C. Pantoea stewartii subsp. stewartii: Lessons learned from a xylem-dwelling pathogen of sweet corn. Mol. Plant Pathol. 2011, 12, 628–637. [Google Scholar] [CrossRef]
- Bartholomew, H.P.; Reynoso, G.; Thomas Brandi, J.; Mullins Chase, M.; Smith, C.; Gentzel, I.N.; Giese, L.A.; Mackey, D.; Stevens, A.M. The Transcription factor Lrp of Pantoea stewartii subsp. stewartii controls capsule production, motility, and virulence important for in planta growth. Front. Microbiol. 2022, 12, 4229. [Google Scholar] [CrossRef]
- Malafaia, C.B.; Jardelino, A.C.S.; Silva, A.G.; de Souza, E.B.; Macedo, A.J.; Correia, M.T.D.S.; Silva, M.V. Effects of caatinga plant extracts in planktonic growth and biofilm formation in Ralstonia solanacearum. Microb. Ecol. 2018, 75, 555–561. [Google Scholar] [CrossRef]
- Pau, S.; de Roger, P.-J.; Benoit, D.; Anurag, K.; Núria, S.C.; Marc, V. The Bacterial wilt reservoir host solanum dulcamara shows resistance to Ralstonia solanacearum Infection. Front. Plant Sci. 2021, 12, 755708. [Google Scholar] [CrossRef]
- Corral, J.; Sebastià, P.; Coll, N.S.; Barbé, J.; Aranda, J.; Valls, M. Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. Msphere. 2020, 5, e00740-19. [Google Scholar] [CrossRef]
- Yoshihara, A.; Shimatani, M.; Sakata, M.; Takemura, C.; Senuma, W.; Hikichi, Y.; Kai, K. Quorum sensing inhibition attenuates the virulence of the plant pathogen Ralstonia solanacearum species complex. ACS Chem. Biol. 2020, 15, 3050–3059. [Google Scholar] [CrossRef]
- Eichenlaub, R.; Gartemann, K.H. The Clavibacter michiganensis subspecies: Molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 2011, 49, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Peritore-Galve, F.C.; Miller, C.; Smart, C.D. Characterizing colonization patterns of Clavibacter michiganensis during infection of tolerant wild Solanum species. Phytopathology 2020, 110, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Chalupowicz, L.; Zellermann, E.M.; Fluegel, M.; Dror, O.; Eichenlaub, R.; Gartemann, K.H.; Savidor, A.; Sessa, G.; Iraki, N.; Barash, I.; et al. Colonization and movement of GFP-labeled Clavibacter michiganensis subsp. michiganensis during tomato infection. Phytopathology 2012, 102, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Flügel, M.; Becker, A.; Gartemann, K.H.; Eichenlaub, R. Analysis of the interaction of Clavibacter michiganensis subsp. michiganensis with its host plant tomato by genome-wide expression profiling. J. Biotechnol. 2022, 160, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Sosa, L.M.; Ramírez-Valverde, G.; Martínez-Yáñez, B.; Judith-Hernández, A.; Aranda-Ocampo, S. Response of tomato (Solanum lycopersicum) varieties to Clavibacter michiganensis subsp. michiganensis infection. Rev. Mex. Fitopatol. 2022, 40, 18–39. [Google Scholar] [CrossRef]
- Padmavathi, A.R.; Bakkiyaraj, D.; Pandian, S.K. Biochemical and molecular mechanisms in biofilm formation of plant-associated bacteria. In Biofilms in Plant and Soil Health; Ahmad, I., Husain, F.M., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 195–214. [Google Scholar] [CrossRef]
- Thompson, M.A.; Onyeziri, M.C.; Fuqua, C. Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. In Agrobacterium Biology. Current Topics in Microbiology and Immunology; Gelvin, S., Ed.; Springer: Cham, Switzerland, 2018; Volume 418, pp. 143–184. [Google Scholar] [CrossRef]
- Kumar Junta, M.; Gupta, A.K.; Mahajan, R. Biological control of hairy root (Rhizobium rhizogenes) in apple nurseries through Rhizobium radiobacter antagonists (strain K-84 and native strain UHFBA-218). Biol. Control. 2021, 164, 104762. [Google Scholar] [CrossRef]
- Bourigault, Y.; Rodrigues, S.; Crépin, A.; Chane, A.; Taupin, L.; Bouteiller, M.; Dupont, C.; Merieau, A.; Konto-Ghiorghi, Y.; Boukerb, A.M.; et al. Biocontrol of biofilm formation: Jamming of sessile-associated rhizobial communication by Rhodococcal Quorum-Quenching. Int. J. Mol. Sci. 2021, 22, 8241. [Google Scholar] [CrossRef]
- Rajkumari, J.; Katiyar, P.; Dheeman, S.; Pandey, P.; Maheshwari, D.K. The changing paradigm of rhizobial taxonomy and its systematic growth upto postgenomic technologies. World J. Microbiol. Biotechnol. 2022, 38, 206. [Google Scholar] [CrossRef]
- Tomihama, T.; Nonaka, T.; Nishi, Y.; Arai, K. Environmental control in tea fields to reduce infection by Pseudomonas syringae pv. theae. Phytopathology 2009, 99, 209–216. [Google Scholar] [CrossRef]
- Ueda, A.; Saneoka, H. Caracterización de la capacidad de formar biopelículas por especies de Pseudomonas asociadas a plantas. Curr. Microbiol. 2015, 70, 506–513. [Google Scholar] [CrossRef]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Patyka, V.; Buletsa, N.M.; Pasichnyk, L.A.; Zhitkevich, N.; Kalinichenko, A.; Gnatiuk, T.T.; Butsenko, L.N. Specifics of pesticides effects on the phytopathogenic bacteria. Ecol. Chem. Eng. S. 2016, 23, 311–331. [Google Scholar] [CrossRef]
- Shao, X.; Xie, Y.; Zhang, Y.; Deng, X. Biofilm formation assay in Pseudomonas syringae. Bio. Protoc. 2019, 9, e3237. [Google Scholar] [CrossRef]
- Fishman, M. Signaling Dynamics in Pseudomonas syringae pv. Tomato DC3000. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2018. [Google Scholar]
- Engl, C.; Waite, C.J.; McKenna, J.F.; Bennett, M.H.; Hamann, T.; Buck, M. Chp8, a Diguanylate cyclase from Pseudomonas syringae pv. tomato DC3000, suppresses the pathogen-associated molecular pattern flagellin, increases extracellular polysaccharides, and promotes plant immune evasion. mBio 2014, 5, e01168-14. [Google Scholar] [CrossRef] [PubMed]
- Carezzano, E.; Sotelo, J.; Primo, E.; Reinoso, E.; Paletti Rovey, M.F.; Demo, M.; Giordano, W.; Oliva, M.D.L.M. Inhibitory effect of Thymus vulgaris and Origanum vulgare EO on virulence factors of phytopathogenic Pseudomonas syringae strains. Plant Biol. 2017, 19, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, A.; Pucci, N.; Modesti, V.; Lumia, V.; Latini, A.; Loreti, S.; Pilotti, M. In vitro and in planta screening of compounds for the control of Pseudomonas syringae pv. actinidiae in Actinidia chinensis var. chinensis. Eur. J. Plant Pathol. 2020, 158, 829–848. [Google Scholar] [CrossRef]
- Han, Q.; Feng, L.; Zhang, Y.; Zhang, R.; Wang, G.; Zhang, Y. Effect of Juglone against Pseudomonas syringae pv. actinidiae planktonic growth and biofilm formation. Molecules 2021, 26, 7580. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Sakata, N.; Usuki, G.; Ishiga, T.; Hashimoto, Y.; Ishiga, Y. Multiple virulence factors regulated by AlgU contribute to the pathogenicity of Pseudomonas savastanoi pv. glycinea in soybean. PeerJ 2021, 9, e12405. [Google Scholar] [CrossRef]
- Fei, N.; Ji, W.; Yang, L.; Yu, C.; Qiao, P.; Yan, J.; Guan, W.; Yang, Y.; Zhao, T. Hcp of the Type VI Secretion System (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int. J. Mol. Sci. 2022, 23, 9632. [Google Scholar] [CrossRef]
- Ji, W.; Zhao, M.; Fei, N.; Yang, L.; Qiao, P.; Walcott, R.; Yang, Y.; Zhao, T. Essential Acidovorax citrulli virulence gene hrpE activates host immune response against pathogen. Int. J. Mol. Sci. 2022, 23, 9144. [Google Scholar] [CrossRef]
- Tambong, J.T. Bacterial Pathogens of Wheat: Symptoms, Distribution, Identification, and Taxonomy. In Wheat; Rahman Ansari, M., Ed.; IntechOpen: Cambridge, MA, USA, 2022. [Google Scholar] [CrossRef]
- Gimranov, E.; Santos, J.D.N.; Vitorino, I.; Martín, J.; Reyes, F.; Moura, L.; Tavares, F.; Santos, C.; Mariz-Ponte, N.; Lage, O.M. Marine bacterial activity against phytopathogenic Pseudomonas show high efficiency of planctomycetes extracts. Eur. J. Plant Pathol. 2022, 162, 843–854. [Google Scholar] [CrossRef]
- Farias, G.A.; Olmedilla, A.; Gallegos, M.T. Visualization and characterization of Pseudomonas syringae pv. tomato DC 3000 pellicles. Microb. Biotechnol. 2019, 12, 688–702. [Google Scholar] [CrossRef]
- Ni, P.; Wang, L.; Deng, B.; Jiu, S.; Ma, C.; Zhang, C.; Almeida, A.; Wang, D.; Xu, W.; Wang, S. Combined application of bacteriophages and carvacrol in the control of Pseudomonas syringae pv. actinidiae planktonic and biofilm forms. Microorganisms 2020, 8, 837. [Google Scholar] [CrossRef]
- Manoharan, B.; Neale, H.C.; Hancock, J.T.; Jackson, R.W.; Arnold, D.L. The Identification of genes important in Pseudomonas syringae pv. phaseolicola plant colonisation using in vitro screening of transposon libraries. PLoS ONE 2015, 10, e0137355. [Google Scholar] [CrossRef]
- Carezzano, M.E.; Paletti Rovey, M.F.; Sotelo, J.P.; Giordano, M.; Bogino, P.; Oliva, M.d.l.M.; Giordano, W. Inhibitory potential of Thymus vulgaris essential oil against growth, biofilm formation, swarming, and swimming in Pseudomonas syringae isolates. Processes 2023, 11, 933. [Google Scholar] [CrossRef]
- Picchi, S.C.; Takita, M.A.; Coletta-Filho, H.D.; Machado, M.A.; de Souza, A.A. N-acetylcysteine interferes with the biofilm formation, motility and epiphytic behaviour of Xanthomonas citri subsp. citri. Plant Pathol. 2016, 65, 561–569. [Google Scholar] [CrossRef]
- Osdaghi, E. Occurrence of common bacterial blight on mungbean (Vigna radiata) in Iran caused by Xanthomonas axonopodis pv. phaseoli. New Dis. Rep. 2014, 30, 2044-0588. [Google Scholar] [CrossRef]
- Hailu, N.; Fininsa, C.; Tana, T.; Mamo, G. Effects of temperature and moisture on growth of common bean and its resistance reaction against common bacterial blight (Xanthomonas axonopodis pv. phaseoli strains). J. Plant Pathol. Microb. 2017, 8, 1000419. [Google Scholar]
- Qi, P.Y.; Zhang, T.H.; Feng, Y.M.; Wang, M.W.; Shao, W.B.; Zeng, D.; Jin, L.H.; Wang, P.Y.; Zhou, X.; Yang, S. Exploring an innovative strategy for suppressing bacterial plant disease: Excavated novel isopropanolamine-Tailored pterostilbene derivatives as potential antibiofilm agents. J. Agric. Food Chem. 2022, 70, 4899–4911. [Google Scholar] [CrossRef] [PubMed]
- Papaianni, M.; Paris, D.; Woo, S.L.; Fulgione, A.; Rigano, M.M.; Parrilli, E.; Tutino, M.L.; Marra, R.; Manganiello, G.; Casillo, A.; et al. Plant Dynamic metabolic response to bacteriophage treatment after Xanthomonas campestris pv. campestris infection. Front. Microbiol. 2020, 11, 732. [Google Scholar] [CrossRef] [PubMed]
- Burbank, L.; Mohammadi, M.; Roper, M.C. Siderophore-mediated iron acquisition influences motility and is required for full virulence of the xylem-dwelling bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 2015, 81, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Naas, H.; Sebaihia, M.; Orfei, B.; Rezzonico, F.; Buonaurio, R.; Moretti, C. Pectobacterium carotovorum subsp. brasiliense and Pectobacterium carotovorum subsp. carotovorum as causal agents of potato soft rot in Algeria. Eur. J. Plant Pathol. 2018, 151, 1027–1034. [Google Scholar] [CrossRef]
- Meng, X.; Chai, A.; Shi, Y.; Xie, X.; Ma, Z.; Li, B. Emergence of bacterial soft rot in cucumber caused by Pectobacterium carotovorum subsp. brasiliense in China. Plant Dis. 2017, 101, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, B.S.; PrasannaKumar, M.K.; Parivallal, P.B.; Pramesh, D.; Banakar, S.N.; Patil, S.S.; Mahesh, H.B. Host range and virulence diversity of Pectobacterium carotovorum subsp. brasiliense strain RDKLR infecting radish in India, and development of a LAMP-based diagnostics. J. Appl. Microbiol. 2022, 132, 4400–4412. [Google Scholar] [CrossRef] [PubMed]
- Heindl, J.E.; Hibbing, M.E.; Xu, J.; Natarajan, R.; Buechlein, A.M.; Fuqua, C. Discrete responses to limitation for iron and manganese in Agrobacterium tumefaciens: Influence on attachment and biofilm formation. J. Bacteriol. 2016, 198, 816–829. [Google Scholar] [CrossRef]
- Li, Y.; Chen, X.; Xu, X.; Yu, C.; Liu, Y.; Jiang, N.; Li, J.; Luo, L. Deletion of pbpC enhances bacterial pathogenicity on tomato by affecting biofilm formation, exopolysaccharides production, and exoenzyme activities in Clavibacter michiganensis. Int. J. Mol. Sci. 2023, 24, 5324. [Google Scholar] [CrossRef]
- Janissen, R.; Murillo, D.M.; Niza, B.; Sahoo, P.K.; Nobrega, M.M.; Cesar, C.L.; Temperini, M.L.A.; Carvalho, H.F.; de Souza, A.A.; Cotta, M.A. Spatiotemporal distribution of different extracellular polymeric substances and filamentation mediate Xylella fastidiosa adhesion and biofilm formation. Sci. Rep. 2015, 5, 9856. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schachterle, J.K.; Sundin, G.W. Orchestration of virulence factor expression and modulation of biofilm dispersal in Erwinia amylovora through activation of the Hfq-dependent small RNA RprA. Mol. Plant Pathol. 2021, 22, 255–270. [Google Scholar] [CrossRef]
- Fontana, R.; Macchi, G.; Caproni, A.; Sicurella, M.; Buratto, M.; Salvatori, F.; Pappadà, M.; Manfredini, S.; Baldisserotto, A.; Marconi, P. Control of Erwinia amylovora growth by Moringa oleifera leaf extracts: In vitro and in planta effects. Plants 2022, 11, 957. [Google Scholar] [CrossRef] [PubMed]
- Rahimi-Midani, A.; Kim, J.O.; Kim, J.H.; Lim, J.; Ryu, J.G.; Kim, M.K.; Choi, T.J. Potential use of newly isolated bacteriophage as a biocontrol against Acidovorax citrulli. Arch. Microbiol. 2020, 202, 377–389. [Google Scholar] [CrossRef] [PubMed]
| Species (Infectious Agent) | Host (Sensitive Plant) | Disease | Symptoms | References | |
|---|---|---|---|---|---|
| Pseudomonas syringae | pv. syringae | Wheat | Bacterial leaf blight | Water-soaked lesions that turn gray-green, leaf necrosis | [154] |
| pv. syringae | Tomato, cereals, citrus plants, kiwifruit | Spot, speck, and bacterial blight | Foliarand stem necrotic lesions; basal stem and root rot | [155] | |
| pv. atrofaciens | Wheat | Basal glume rot | Dull brownish-blackish discoloration on the lower part of the glume | [154] | |
| pv. tomato | Tomato | Bacterial speck | Chlorosis and necrotic lesions | [156] | |
| pv. actinidiae | Kiwi | Bacterial bleeding canker | Brown leaf spots with chlorotic haloes, fruit specks and scabs, brown discoloration of buds, and cankers with exudates on trunks and twigs | [157] | |
| pv. phaseolicola | Bean | Halo blight | Necrotic lesions on the leaf | [158] | |
| Pseudomonas savastanoi pv. glycinea | Soybean | Bacterial leaf blight | Circular necrotic lesions on leaves surrounded by a chlorotic halo | [151,159] | |
| Xanthomonas citri subsp. Citri | Citrus plants | Citrus canker | Erumpent lesions on fruit, foliage, and young stems | [160] | |
| Xanthomonas axonopodis pv. phaseoli | Bean | Common bacterial blight | Dark green water-soaked lesions, necrotic symptoms on the margins of leaves | [161,162] | |
| Xanthomonas oryzae pv. oryzae | Rice | Bacterial leaf blight | Tannish gray-white lesions along the veins | [51,163] | |
| Xanthomonas campestris pv. campestris | Cruciferous plants | Black rot | V-shaped necrotic lesions on the foliar margins and blackened veins | [164] | |
| Xanthomonas translucens pv. undulosa | Wheat | Bacterial streak and black chaff disease | Water-soaked necrotic streaks which eventually change into translucent lesions | [154] | |
| Pantoea stewartii subsp. stewartii | Corn | Stewart’s wilt, severe seedling wilt | Water-soaked lesions, wilting in young seedlings | [165] | |
| Pectobacterium carotovorum subsp. carotovorum | Potato | Soft rot | Severe bacterial tuber soft rot | [166] | |
| Pectobacterium carotovorum subsp. brasiliense | Potato, tomato, cucumber, radish | Soft rot | Severe and typical bacterial soft rot, water-soaked and macerated tissues | [167,168] | |
| Agrobacterium tumefaciens | Dicotyledonous plants | Crown gall | Tumors | [169] | |
| Rhizobium rhizogenes | Tomato, cucumber, apple | Hairy root | Smaller root structures spring out at right angles from the main root. In the aerial form, tumors or knots (woolly-knots) appear in the limbs | [137,138] | |
| Clavibacter michiganensis | Tomato | Bacterial canker, wilt disease | Deterioration of the internal vascular tissues, stem cankers, foliar chlorosis, unilateral wilt, marginal leaf necrosis, fruits with localized bird’s-eye spots | [131,170] | |
| Xylella fastidiosa | Citrus plants grape, coffee, almond, olives, peach, blueberry, among others | Pierce’s disease, leaf scorch, and citrus-variegated chlorosis | Leaf chlorosis, marginal scorching, and/or dwarfing, depending on the host | [5,171] | |
| Erwinia amylovora | Apple, pear | Fire blight | “Shepherd’s crook” of the twigs and a yellowish bacterial exudate on the infected tissues. Infection of leaves at shoot tips, wilting of leaves, cankers | [172,173] | |
| Ralstonia solanacearum | Tomato, brinjal, tobacco, potato, banana | Bacterial wilt | Rolling of leaves, chlorosis, and necrosis | [109] | |
| Dickeya dadantii | Sweet potato | Bacterial stem and root rot | Maceration of plant tissues | [117] | |
| Acidovorax citrulli | Melon, watermelon, pumpkin | Bacterial fruit blotch | Water-soaked seedlings and light brown-reddish lesions on the leaves, small water-soaked regions on the fruit surface | [126,174] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carezzano, M.E.; Paletti Rovey, M.F.; Cappellari, L.d.R.; Gallarato, L.A.; Bogino, P.; Oliva, M.d.l.M.; Giordano, W. Biofilm-Forming Ability of Phytopathogenic Bacteria: A Review of its Involvement in Plant Stress. Plants 2023, 12, 2207. https://doi.org/10.3390/plants12112207
Carezzano ME, Paletti Rovey MF, Cappellari LdR, Gallarato LA, Bogino P, Oliva MdlM, Giordano W. Biofilm-Forming Ability of Phytopathogenic Bacteria: A Review of its Involvement in Plant Stress. Plants. 2023; 12(11):2207. https://doi.org/10.3390/plants12112207
Chicago/Turabian StyleCarezzano, María Evangelina, María Fernanda Paletti Rovey, Lorena del Rosario Cappellari, Lucas Antonio Gallarato, Pablo Bogino, María de las Mercedes Oliva, and Walter Giordano. 2023. "Biofilm-Forming Ability of Phytopathogenic Bacteria: A Review of its Involvement in Plant Stress" Plants 12, no. 11: 2207. https://doi.org/10.3390/plants12112207
APA StyleCarezzano, M. E., Paletti Rovey, M. F., Cappellari, L. d. R., Gallarato, L. A., Bogino, P., Oliva, M. d. l. M., & Giordano, W. (2023). Biofilm-Forming Ability of Phytopathogenic Bacteria: A Review of its Involvement in Plant Stress. Plants, 12(11), 2207. https://doi.org/10.3390/plants12112207






