Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses
Abstract
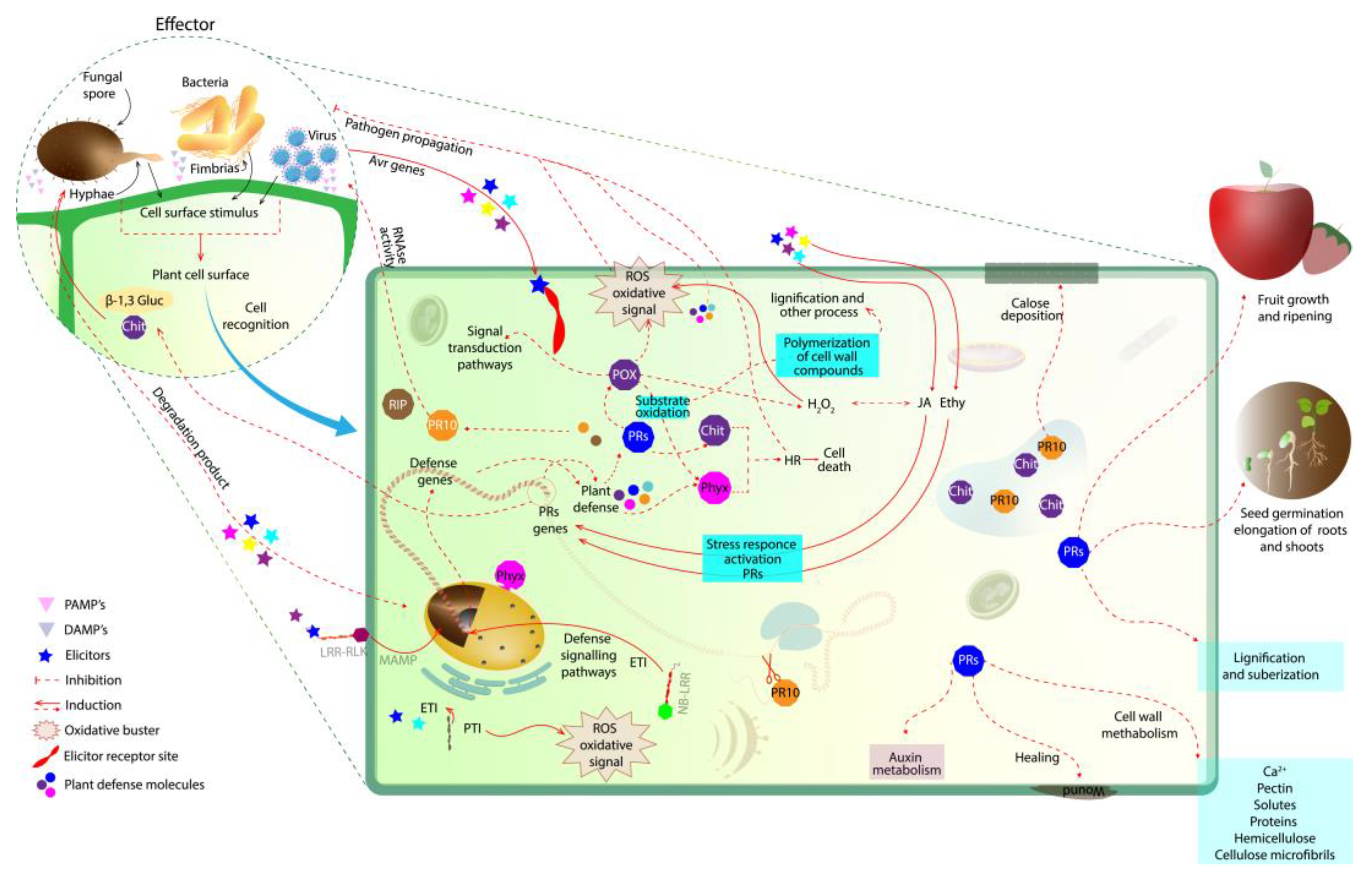
1. Introduction
2. PR Proteins with Enzymatic Action and the Role in Plant Defense Activation
2.1. PR-2 and PR-3 Families: β-1,3 Glucanases and Chitinases
2.2. PR-9 Family: Peroxidases
2.3. PR-10 Family: Ribonucleases
3. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, P.; Lu, Y.-J.; Chen, H.; Day, B. The Lifecycle of the Plant Immune System. CRC Crit. Rev. Plant Sci. 2020, 39, 72–100. [Google Scholar] [CrossRef] [PubMed]
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of Plant Defense System in Response to Microbial Interactions. Front. Microbiol. 2020, 11, 1298. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.-M.; Iswanto, A.B.B.; Son, G.H.; Kim, S.H. Recent advances in effector-triggered immunity in plants: New pieces in the puzzle create a different paradigm. Int. J. Mol. Sci. 2021, 22, 4709. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.-F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Freeman, B.; Beattie, G. An overview of plant defenses against pathogens and herbivores. Overv. Plant Def. 2008. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Souza, T.P.; Dias, R.O.; Silva-Filho, M.C. Defense-related proteins involved in sugarcane responses to biotic stress. Genet. Mol. Biol. 2017, 40, 360–372. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant pathogenesis-related proteins PR-10 and PR-14 as components of innate immunity system and ubiquitous allergens. Curr. Med. Chem. 2017, 24, 1772–1787. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Yildiz, S.; Filiz, E. Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics 2020, 112, 4089–4099. [Google Scholar] [CrossRef]
- Pečenková, T.; Pejchar, P.; Moravec, T.; Drs, M.; Haluška, S.; Šantrůček, J.; Potocká, A.; Žárský, V.; Potocký, M. Immunity functions of Arabidopsis pathogenesis-related 1 are coupled but not confined to its C-terminus processing and trafficking. Mol. Plant Pathol. 2022, 23, 664–678. [Google Scholar] [CrossRef]
- Almeida-Silva, F.; Venancio, T.M. Pathogenesis-related protein 1 (PR-1) genes in soybean: Genome-wide identification, structural analysis and expression profiling under multiple biotic and abiotic stresses. Gene 2022, 809, 146013. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Edwards, M.C. Molecular characterization and functional analysis of PR-1-like proteins identified from the wheat head blight fungus Fusarium graminearum. Phytopathology 2018, 108, 510–520. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Shaw, J.; Yu, C.; Makhotenko, A.V.; Makarova, S.S.; Love, A.J.; Kalinina, N.O.; MacFarlane, S.; Chen, J.; Taliansky, M.E. Interaction of a plant virus protein with the signature Cajal body protein coilin facilitates salicylic acid-mediated plant defence responses. New Phytol. 2019, 224, 439–453. [Google Scholar] [CrossRef]
- Sung, Y.C.; Outram, M.A.; Breen, S.; Wang, C.; Dagvadorj, B.; Winterberg, B.; Kobe, B.; Williams, S.J.; Solomon, P.S. PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 2021, 229, 3467–3480. [Google Scholar] [CrossRef]
- Anisimova, O.K.; Shchennikova, A.V.; Kochieva, E.Z.; Filyushin, M.A. Pathogenesis-Related Genes of PR1, PR2, PR4, and PR5 Families Are Involved in the Response to Fusarium Infection in Garlic (Allium sativum L.). Int. J. Mol. Sci. 2021, 22, 6688. [Google Scholar] [CrossRef]
- Boccardo, N.A.; Segretin, M.E.; Hernandez, I.; Mirkin, F.G.; Chacón, O.; Lopez, Y.; Borrás-Hidalgo, O.; Bravo-Almonacid, F.F. Expression of pathogenesis-related proteins in transplastomic tobacco plants confers resistance to filamentous pathogens under field trials. Sci. Rep. 2019, 9, 2791. [Google Scholar] [CrossRef]
- Enoki, S.; Suzuki, S. Pathogenesis-related proteins in grape. Grape Wine Biotechnol. 2016, 43, 46–49. [Google Scholar] [CrossRef]
- Santos, C.; Nogueira, F.C.S.; Domont, G.B.; Fontes, W.; Prado, G.S.; Habibi, P.; Santos, V.O.; Oliveira-Neto, O.B.; Grossi-de-Sá, M.F.; Jorrín-Novo, J.V.; et al. Proteomic analysis and functional validation of a Brassica oleracea endochitinase involved in resistance to Xanthomonas campestris. Front. Plant Sci. 2019, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, F.; Liang, F.; Zhang, Y.; Ma, L.; Wang, H.; Liu, D. Functional analysis of a pathogenesis-related thaumatin-like protein gene TaLr35PR5 from wheat induced by leaf rust fungus. BMC Plant Biol. 2018, 18, 76. [Google Scholar] [CrossRef] [PubMed]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A cationic protein leads to improve biotic and abiotic stress tolerance in plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef]
- Rodríguez-Sifuentes, L.; Marszalek, J.E.; Chuck-Hernández, C.; Serna-Saldívar, S.O. Legumes protease inhibitors as biopesticides and their defense mechanisms against biotic factors. Int. J. Mol. Sci. 2020, 21, 3322. [Google Scholar] [CrossRef]
- Kaur, A.; Kaur, S.; Kaur, A.; Sarao, N.K.; Sharma, D. Pathogenesis-Related Proteins and Their Transgenic Expression for Developing Disease-Resistant Crops: Strategies Progress and Challenges. In Plant Breeding - New Perspectives; IntechOpen: London, UK, 2022; pp. 6–8. [Google Scholar] [CrossRef]
- Sudisha, J.; Sharathchandra, R.G.; Amruthesh, K.N.; Kumar, A.; Shetty, H.S. Pathogenesis related proteins in plant defense response. In Plant Defence: Biological Control; Springer: Dordrecht, Netherlands, 2012; pp. 379–403. [Google Scholar]
- Irigoyen, M.L.; Garceau, D.C.; Bohorquez-Chaux, A.; Lopez-Lavalle, L.A.B.; Perez-Fons, L.; Fraser, P.D.; Walling, L.L. Genome-wide analyses of cassava Pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid. BMC Genom. 2020, 21, 93. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Kim, H.S.; Park, S.-C.; Ji, C.Y.; Yang, J.W.; Lee, H.U.; Kwak, S.-S. Overexpression of swpa4 peroxidase enhances tolerance to hydrogen peroxide and high salinity-mediated oxidative stress in transgenic sweetpotato plants. Plant Biotechnol. Rep. 2020, 14, 301–307. [Google Scholar] [CrossRef]
- Sellami, K.; Couvert, A.; Nasrallah, N.; Maachi, R.; Abouseoud, M.; Amrane, A. Peroxidase enzymes as green catalysts for bioremediation and biotechnological applications: A review. Sci. Total Environ. 2022, 806, 150500. [Google Scholar] [CrossRef]
- Wu, J.; Kim, S.G.; Kang, K.Y.; Kim, J.-G.; Park, S.-R.; Gupta, R.; Kim, Y.H.; Wang, Y.; Kim, S.T. Overexpression of a pathogenesis-related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol. J. 2016, 32, 552. [Google Scholar] [CrossRef]
- Mahmoud, F.; Bondok, A.; Khalifa, W.; Abou Abbas, F. Enhancing Tomato Plant Resistance Against Tobacco Mosaic Virus Using Riboflavin. Arab Univ. J. Agric. Sci. 2020, 28, 1257–1270. [Google Scholar] [CrossRef]
- Peng, Q.; Su, Y.; Ling, H.; Ahmad, W.; Gao, S.; Guo, J.; Que, Y.; Xu, L. A sugarcane pathogenesis-related protein, ScPR10, plays a positive role in defense responses under Sporisorium scitamineum, SrMV, SA, and MeJA stresses. Plant Cell Rep. 2017, 36, 1427–1440. [Google Scholar] [CrossRef]
- Franco, O.L.; Murad, A.M.; Leite, J.R.; Mendes, P.A.M.; Prates, M.V.; Bloch Jr, C. Identification of a cowpea γ-thionin with bactericidal activity. FEBS J. 2006, 273, 3489–3497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Höng, K.; Austerlitz, T.; Bohlmann, T.; Bohlmann, H. The thionin family of antimicrobial peptides. PLoS ONE 2021, 16, e0254549. [Google Scholar] [CrossRef]
- Scheurer, S.; Schülke, S. Interaction of Non-Specific Lipid-Transfer Proteins With Plant-Derived Lipids and Its Impact on Allergic Sensitization. Front. Immunol. 2018, 9, 1389. [Google Scholar] [CrossRef]
- Wang, C.; Gao, H.; Chu, Z.; Ji, C.; Xu, Y.; Cao, W.; Zhou, S.; Song, Y.; Liu, H.; Zhu, C. A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato. Mol. Plant Pathol. 2021, 22, 48–63. [Google Scholar] [CrossRef]
- Fleury, C.; Gracy, J.; Gautier, M.-F.; Pons, J.-L.; Dufayard, J.-F.; Labesse, G.; Ruiz, M.; De Lamotte, F. Comprehensive classification of the plant non-specific lipid transfer protein superfamily towards its sequence–structure–function analysis. PeerJ 2019, 7, e7504. [Google Scholar] [CrossRef]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. Physico-chemical characterization and topological analysis of pathogenesis-related proteins from Arabidopsis thaliana and Oryza sativa using in-silico approaches. PLoS ONE 2020, 15, e0239836. [Google Scholar] [CrossRef]
- Farvardin, A.; González-Hernández, A.I.; Llorens, E.; García-Agustín, P.; Scalschi, L.; Vicedo, B. The apoplast: A key player in plant survival. Antioxidants 2020, 9, 604. [Google Scholar] [CrossRef]
- Okushima, Y.; Koizumi, N.; Kusano, T.; Sano, H. Secreted proteins of tobacco cultured BY2 cells: Identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 2000, 42, 479–488. [Google Scholar] [CrossRef]
- Custers, J.H.H.V.; Harrison, S.J.; Sela-Buurlage, M.B.; Van Deventer, E.; Lageweg, W.; Howe, P.W.; Van Der Meijs, P.J.; Ponstein, A.S.; Simons, B.H.; Melchers, L.S. Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 2004, 39, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Sooriyaarachchi, S.; Jaber, E.; Covarrubias, A.S.; Ubhayasekera, W.; Asiegbu, F.O.; Mowbray, S.L. Expression and β-glucan binding properties of Scots pine (Pinus sylvestris L.) antimicrobial protein (Sp-AMP). Plant Mol. Biol. 2011, 77, 33–45. [Google Scholar] [CrossRef]
- Jain, D.; Khurana, J.P. Role of Pathogenesis-Related (PR) Proteins in Plant Defense Mechanism. In Molecular Aspects of Plant-Pathogen Interaction; Singh, A., Singh, I.K., Eds.; Springer: Singapore, 2018; pp. 265–281. [Google Scholar]
- Zribi, I.; Ghorbel, M.; Brini, F. Pathogenesis Related Proteins (PRs): From Cellular Mechanisms to Plant Defense. Curr. Protein Pept. Sci. 2021, 22, 396–412. [Google Scholar] [CrossRef]
- Ali, Z.; Serag, M.; Demirer, G.; Torre, B.; di Fabrizio, E.; Landry, M.; Habuchi, S.; Mahfouz, M. The DNA–carbon nanotube binding mode determines the efficiency of carbon nanotube-mediated DNA delivery to intact plants. ACS Appl. Nano Mater. 2021, 5, 4663–4676. [Google Scholar] [CrossRef]
- Héloir, M.-C.; Adrian, M.; Brulé, D.; Claverie, J.; Cordelier, S.; Daire, X.; Dorey, S.; Gauthier, A.; Lemaître-Guillier, C.; Negrel, J.; et al. Recognition of Elicitors in Grapevine: From MAMP and DAMP Perception to Induced Resistance. Front. Plant Sci. 2019, 10, 1117. [Google Scholar] [CrossRef]
- Jamiołkowska, A. Natural Compounds as Elicitors of Plant Resistance Against Diseases and New Biocontrol Strategies. Agronomy 2020, 10, 173. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Wang, X.; Zhong, L.; Shi, G.; Xu, Y.; Li, Y.; Li, R.; Huang, Y.; Ye, X.; et al. A novel β-1,3-glucanase Gns6 from rice possesses antifungal activity against Magnaporthe oryzae. J. Plant Physiol. 2021, 265, 153493. [Google Scholar] [CrossRef]
- Stintzi, A.; Heitz, T.; Prasad, V.; Wiedemann-Merdinoglu, S.; Kauffmann, S.; Geoffroy, P.; Legrand, M.; Fritig, B. Plant ‘pathogenesis-related’proteins and their role in defense against pathogens. Biochimie 1993, 75, 687–706. [Google Scholar] [CrossRef]
- Ding, Y.; Perez-Ortiz, G.; Peate, J.; Barry, S.M. Redesigning Enzymes for Biocatalysis: Exploiting Structural Understanding for Improved Selectivity. Front. Mol. Biosci. 2022, 9, 684. [Google Scholar] [CrossRef]
- Engqvist, M.K.M.; Rabe, K.S. Applications of protein engineering and directed evolution in plant research. Plant Physiol. 2019, 179, 907–917. [Google Scholar] [CrossRef]
- Talavera-Caro, A.G.; Alva-Sánchez, D.L.; Sosa-Herrera, A.; Sánchez-Muñoz, M.A.; Hernández-De Lira, I.O.; Hernández-Beltran, J.U.; Hernández-Almanza, A.Y.; Balagurusamy, N. Emerging trends and future perspectives on enzyme prospection with reference to food processing. In Value-Addition in Food Products and Processing Through Enzyme Technology; Elsevier Academic Press: London, UK, 2022; pp. 139–151. [Google Scholar] [CrossRef]
- Linton, S.M. The structure and function of cellulase (endo-β-1, 4-glucanase) and hemicellulase (β-1, 3-glucanase and endo-β-1, 4-mannase) enzymes in invertebrates that consume materials ranging from microbes, algae to leaf litter. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2020, 240, 110354. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1, 3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Geelen, D. Callose homeostasis at plasmodesmata: Molecular regulators and developmental relevance. Front. Plant Sci. 2014, 5, 138. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gimenez, G.; Russell, J.; Aubert, M.K.; Fincher, G.B.; Burton, R.A.; Waugh, R.; Tucker, M.R.; Houston, K. Barley grain (1, 3; 1, 4)-β-glucan content: Effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci. Rep. 2019, 9, 17250. [Google Scholar] [CrossRef] [PubMed]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging Roles of β-Glucanases in Plant Development and Adaptative Responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Paniagua, C.; Perry, L.; Benitez-Alfonso, Y. A phylogenetic and transcriptomic study of the β-1, 3-glucanase family in tomato identifies candidate targets for fruit improvement. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wu, Q.; Dou, X.; Wang, Q.; Guan, Z.; Cai, Y.; Liao, X. Isolation of β-1,3-Glucanase-Producing Microorganisms from Poria cocos Cultivation Soil via Molecular Biology. Molecules 2018, 23, 1555. [Google Scholar] [CrossRef]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant–fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Wanke, A.; Malisic, M.; Wawra, S.; Zuccaro, A. Unraveling the sugar code: The role of microbial extracellular glycans in plant–microbe interactions. J. Exp. Bot. 2021, 72, 15–35. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, N.; Wang, K.; Xian, Q.; Dong, J.; Qi, X.; Chen, X. Chitinase Chi 2 Positively Regulates Cucumber Resistance against Fusarium oxysporum f. sp. cucumerinum. Genes 2022, 13, 62. [Google Scholar] [CrossRef]
- Liu, X.; Yu, Y.; Liu, Q.; Deng, S.; Jin, X.; Yin, Y.; Guo, J.; Li, N.; Liu, Y.; Han, S.; et al. A Na2CO3-Responsive Chitinase Gene From Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Xin, Y.; Wang, D.; Han, S.; Li, S.; Gong, N.; Fan, Y.; Ji, X. Characterization of the chitinase gene family in mulberry (Morus notabilis) and MnChi18 involved in resistance to Botrytis cinerea. Genes 2021, 13, 98. [Google Scholar] [CrossRef]
- Ali, M.; Li, Q.H.; Zou, T.; Wei, A.M.; Gombojab, G.; Lu, G.; Gong, Z.H. Chitinase Gene Positively Regulates Hypersensitive and Defense Responses of Pepper to Colletotrichum acutatum Infection. Int. J. Mol. Sci. 2020, 21, 6624. [Google Scholar] [CrossRef]
- Franzener, G.; Schwan-Estrada, K.R.F.; Moura, G.S.; Kuhn, O.J.; Stangarlin, J.R. Induction of defense enzymes and control of anthracnose in cucumber by Corymbia citriodora aqueous extract. Summa Phytopathol. 2018, 44, 10–16. [Google Scholar] [CrossRef]
- Oliveira, M.D.M.; Varanda, C.M.R.; Félix, M.R.F. Induced resistance during the interaction pathogen x plant and the use of resistance inducers. Phytochem. Lett. 2016, 15, 152–158. [Google Scholar] [CrossRef]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Sorokan, A.V.; Burhanova, G.F.; Maksimov, I.V. Anionic peroxidase-mediated oxidative burst requirement for jasmonic acid-dependent Solanum tuberosum defence against Phytophthora infestans. Plant Pathol. 2018, 67, 349–357. [Google Scholar] [CrossRef]
- Bela, K.; Riyazuddin, R.; Csiszár, J. Plant Glutathione Peroxidases: Non-Heme Peroxidases with Large Functional Flexibility as a Core Component of ROS-Processing Mechanisms and Signalling. Antioxidants 2022, 11, 1624. [Google Scholar] [CrossRef]
- Twala, P.P.; Mitema, A.; Baburam, C.; Feto, N.A. Breakthroughs in the discovery and use of different peroxidase isoforms of microbial origin. AIMS Microbiol. 2020, 6, 330–349. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef]
- Blaschek, L.; Pesquet, E. Phenoloxidases in plants—How structural diversity enables functional specificity. Front. Plant Sci. 2021, 12, 754601. [Google Scholar] [CrossRef] [PubMed]
- Campos, Â.D.; Ferreira, A.G.; Hampe, M.M.V.; Antunes, I.F.; Brancão, N.; Silveira, E.P.d.; Osório, V.A.; Augustin, E. Peroxidase and polyphenol oxidase activity in bean anthracnose resistance. Pesqui. Agropecu. Bras. 2004, 39, 637–643. [Google Scholar] [CrossRef]
- Basumatary, D.; Yadav, H.S.; Yadav, M. The role of peroxidases in the bioremediation of organic pollutants. Nat. Prod. J. 2023, 13, 60–77. [Google Scholar] [CrossRef]
- Kurnik, K.; Treder, K.; Skorupa-Kłaput, M.; Tretyn, A.; Tyburski, J. Removal of phenol from synthetic and industrial wastewater by potato pulp peroxidases. Water Air Soil Pollut. 2015, 226, 254. [Google Scholar] [CrossRef]
- Lakshmi, S.; Shashidhara, G.M.; Madhu, G.M.; Muthappa, R.; Vivek, H.K.; Nagendra Prasad, M.N. Characterization of peroxidase enzyme and detoxification of phenols using peroxidase enzyme obtained from Zea mays L waste. Appl. Water Sci. 2018, 8, 207. [Google Scholar] [CrossRef]
- Zechmann, B. Subcellular Roles of Glutathione in Mediating Plant Defense during Biotic Stress. Plants 2020, 9, 1067. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Hossain, M.S.; Mahmud, J.A.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated Actions of Glyoxalase and Antioxidant Defense Systems in Conferring Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef]
- Kámán-Tóth, E.; Dankó, T.; Gullner, G.; Bozsó, Z.; Palkovics, L.; Pogány, M. Contribution of cell wall peroxidase-and NADPH oxidase-derived reactive oxygen species to Alternaria brassicicola-induced oxidative burst in Arabidopsis. Mol. Plant Pathol. 2019, 20, 485–499. [Google Scholar] [CrossRef]
- Zhao, L.; Phuong, L.T.; Luan, M.T.; Fitrianti, A.N.; Matsui, H.; Nakagami, H.; Noutoshi, Y.; Yamamoto, M.; Ichinose, Y.; Shiraishi, T. A class III peroxidase PRX34 is a component of disease resistance in Arabidopsis. J. Gen. Plant Pathol. 2019, 85, 405–412. [Google Scholar] [CrossRef]
- dos Santos, C.; Carmo, L.S.T.; Távora, F.T.P.K.; Lima, R.F.C.; Da Nobrega Mendes, P.; Labuto, L.B.D.; De Sá, M.E.L.; Grossi-De-Sa, M.F.; Mehta, A. Overexpression of cotton genes GhDIR4 and GhPRXIIB in Arabidopsis thaliana improves plant resistance to root-knot nematode (Meloidogyne incognita) infection. 3 Biotech 2022, 12, 211. [Google Scholar] [CrossRef]
- Do, J.H.; Park, S.Y.; Park, S.H.; Kim, H.M.; Ma, S.H.; Mai, T.D.; Shim, J.S.; Joung, Y.H. Development of a genome-edited tomato with high ascorbate content during later stage of fruit ripening through mutation of SlAPX4. Front. Plant Sci. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef]
- Wu, B.; Wang, B. Comparative analysis of ascorbate peroxidases (APXs) from selected plants with a special focus on Oryza sativa employing public databases. PLoS ONE 2019, 14, e0226543. [Google Scholar] [CrossRef]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-a potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Chin, D.-C.; Kumar, R.S.; Suen, C.-S.; Chien, C.-Y.; Hwang, M.-J.; Hsu, C.-H.; Xuhan, X.; Lai, Z.X.; Yeh, K.-W. Plant cytosolic ascorbate peroxidase with dual catalytic activity modulates abiotic stress tolerances. Iscience 2019, 16, 31–49. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Poór, P.; Szepesi, Á.; Horváth, E.; Rigó, G.; Szabados, L.; Fehér, A.; Csiszár, J. Crosstalk between the Arabidopsis glutathione peroxidase-like 5 Isoenzyme (AtGPXL5) and ethylene. Int. J. Mol. Sci. 2022, 23, 5749. [Google Scholar] [CrossRef]
- Li, Q.; Qin, X.; Qi, J.; Dou, W.; Dunand, C.; Chen, S.; He, Y. CsPrx25, a class III peroxidase in Citrus sinensis, confers resistance to citrus bacterial canker through the maintenance of ROS homeostasis and cell wall lignification. Hortic. Res. 2020, 7, 192. [Google Scholar] [CrossRef]
- Kuo, E.Y.; Cai, M.-S.; Lee, T.-M. Ascorbate peroxidase 4 plays a role in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Sci. Rep. 2020, 10, 13287. [Google Scholar] [CrossRef]
- Aglas, L.; Soh, W.T.; Kraiem, A.; Wenger, M.; Brandstetter, H.; Ferreira, F. Ligand binding of PR-10 proteins with a particular focus on the Bet v 1 allergen family. Curr. Allergy Asthma Rep. 2020, 20, 25. [Google Scholar] [CrossRef]
- Ghorbel, M.; Zribi, I.; Missaoui, K.; Drira-Fakhfekh, M.; Azzouzi, B.; Brini, F. Differential regulation of the durum wheat Pathogenesis-related protein (PR1) by Calmodulin TdCaM1. 3 protein. Mol. Biol. Rep. 2021, 48, 347–362. [Google Scholar] [CrossRef]
- Longsaward, R.; Pengnoo, A.; Kongsawadworakul, P.; Viboonjun, U. A novel rubber tree PR-10 protein involved in host-defense response against the white root rot fungus Rigidoporus microporus. BMC Plant Biol. 2023, 23, 157. [Google Scholar] [CrossRef] [PubMed]
- Zarattini, M.; Launay, A.; Farjad, M.; Wénès, E.; Taconnat, L.; Boutet, S.; Bernacchia, G.; Fagard, M. The bile acid deoxycholate elicits defences in Arabidopsis and reduces bacterial infection. Mol. Plant Pathol. 2017, 18, 540–554. [Google Scholar] [CrossRef] [PubMed]
- Besbes, F.; Habegger, R.; Schwab, W. Induction of PR-10 genes and metabolites in strawberry plants in response to Verticillium dahliae infection. BMC Plant Biol. 2019, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Genzel, F.; Franken, P.; Witzel, K.; Grosch, R. Systemic induction of salicylic acid-related plant defences in potato in response to Rhizoctonia solani AG 3 PT. Plant Pathol. 2018, 67, 337–348. [Google Scholar] [CrossRef]
- Besbes, F.; Franz-Oberdorf, K.; Schwab, W. Phosphorylation-dependent ribonuclease activity of Fra a 1 proteins. J. Plant Physiol. 2019, 233, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Paek, K.-H. Plant RNA binding proteins for control of RNA virus infection. Front. Physiol. 2013, 4, 397. [Google Scholar] [CrossRef]
- Islam, M.M.; Qi, S.; Zhang, S.; Amin, B.; Yadav, V.; El-Sappah, A.H.; Zhang, F.; Liang, Y. Genome-Wide Identification and Functions against Tomato Spotted Wilt Tospovirus of PR-10 in Solanum lycopersicum. Int. J. Mol. Sci. 2022, 23, 1502. [Google Scholar] [CrossRef]
- Wieczorek, P.; Wrzesińska, B.; Frąckowiak, P.; Przybylska, A.; Obrępalska-Stęplowska, A. Contribution of Tomato torrado virus Vp26 coat protein subunit to systemic necrosis induction and virus infectivity in Solanum lycopersicum. Virol. J. 2019, 16, 9. [Google Scholar] [CrossRef]
- Sinha, R.K.; Verma, S.S.; Rastogi, A. Role of Pathogen-Related Protein 10 (PR 10) under abiotic and biotic stresses in plants. Phyton 2020, 89, 167. [Google Scholar] [CrossRef]
- McBride, J.K.; Cheng, H.; Maleki, S.J.; Hurlburt, B.K. Purification and characterization of pathogenesis related class 10 panallergens. Foods 2019, 8, 609. [Google Scholar] [CrossRef]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the cytosolic interaction between pathogenesis-related protein 10 and leucine-rich repear protein 1 for cell death and defense signaling in pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [CrossRef]
- Lee, O.R.; Pulla, R.K.; Kim, Y.J.; Balusamy, S.R.; Yang, D.C. Expression and stress tolerance of PR10 genes from Panax ginseng C. A. Meyer. Mol. Biol. Rep. 2012, 39, 2365–2374. [Google Scholar] [CrossRef]
- Andrade, L.B.d.S.; Oliveira, A.S.; Ribeiro, J.K.C.; Kiyota, S.; Vasconcelos, I.M.; de Oliveira, J.T.A.; de Sales, M.P. Effects of a novel pathogenesis-related class 10 (PR-10) protein from crotalaria pallida roots with papain inhibitory activity against root-knot nematode Meloidogyne incognita. J. Agric. Food Chem. 2010, 58, 4145–4152. [Google Scholar] [CrossRef]
- Li, B.; Wang, R.; Wang, S.; Zhang, J.; Chang, L. Diversified regulation of cytokinin levels and signaling during Botrytis cinerea infection in Arabidopsis. Front. Plant Sci. 2021, 12, 584042. [Google Scholar] [CrossRef]
- Gao, L. Structure Analysis of a Pathogenesis-Related 10 Protein from Gardenia jasminoides. IOP Conf. Ser. Earth Environ. Sci. 2019, 2019, 042005. [Google Scholar] [CrossRef]
- Wang, W.; Nie, J.; Lv, L.; Gong, W.; Wang, S.; Yang, M.; Xu, L.; Li, M.; Du, H.; Huang, L. A Valsa mali Effector Protein 1 Targets Apple (Malus domestica) Pathogenesis-Related 10 Protein to Promote Virulence. Front. Plant Sci. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Romera, F.J.; García, M.J.; Lucena, C.; Martínez-Medina, A.; Aparicio, M.A.; Ramos, J.; Alcántara, E.; Angulo, M.; Pérez-Vicente, R. Induced Systemic Resistance (ISR) and Fe Deficiency Responses in Dicot Plants. Front. Plant Sci. 2019, 10, 287. [Google Scholar] [CrossRef]
| Family | Properties/Functions | References |
|---|---|---|
| PR-1 (11a, 1b and 1c) |
| [11,12,13,14,15,16,17] |
| PR-2 (Classes: I, II, and III) |
| [15,18,19,20] |
| PR-3; PR-4; PR-8; PR-11 (Classes: I, II, IV, V, VI, and VII) |
| [15,18,20,21] |
| PR-5 |
| [18,22,23] |
| PR-6 |
| [24,25,26] |
| PR-7 |
| [25,26,27] |
| PR-9 |
| [28,29] |
| PR-10 |
| [20,30,31,32] |
| PR-12 |
| [15,18,26] |
| PR-13 (Classes: I, II, III and IV) |
| [26,33,34,35,36] |
| PR-14 |
| [26,37,38,39] |
| PR-15; PR-16 |
| [25,26,40,41] |
| PR-17 |
| [18,25,42] |
| PR-18 |
| [25,43] |
| PR-19 |
| [25,44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, C.; Franco, O.L. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants 2023, 12, 2226. https://doi.org/10.3390/plants12112226
dos Santos C, Franco OL. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants. 2023; 12(11):2226. https://doi.org/10.3390/plants12112226
Chicago/Turabian Styledos Santos, Cristiane, and Octávio Luiz Franco. 2023. "Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses" Plants 12, no. 11: 2226. https://doi.org/10.3390/plants12112226
APA Styledos Santos, C., & Franco, O. L. (2023). Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants, 12(11), 2226. https://doi.org/10.3390/plants12112226







