Effectiveness of R1-nj Anthocyanin Marker in the Identification of In Vivo Induced Maize Haploid Embryos
Abstract
1. Introduction
2. Results
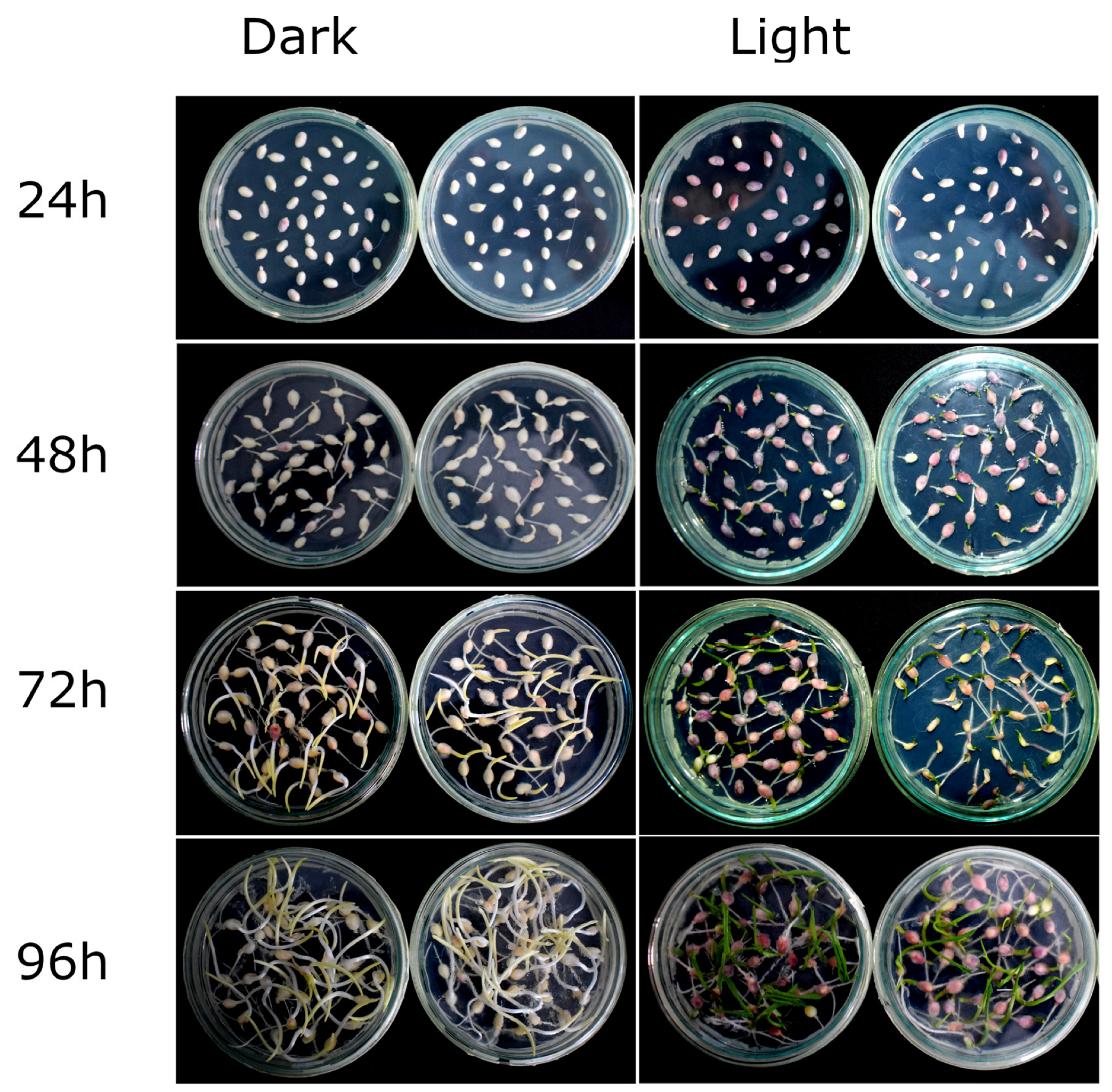
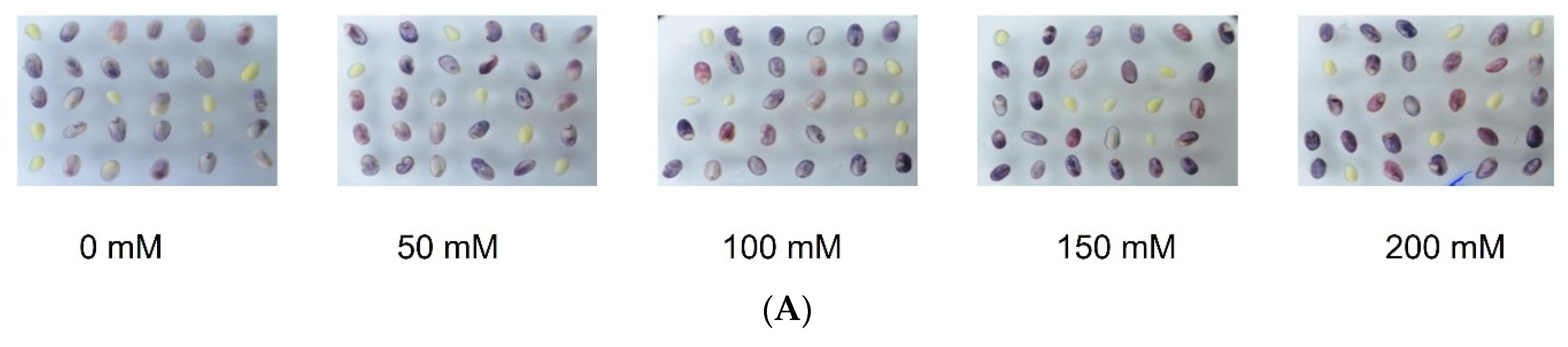
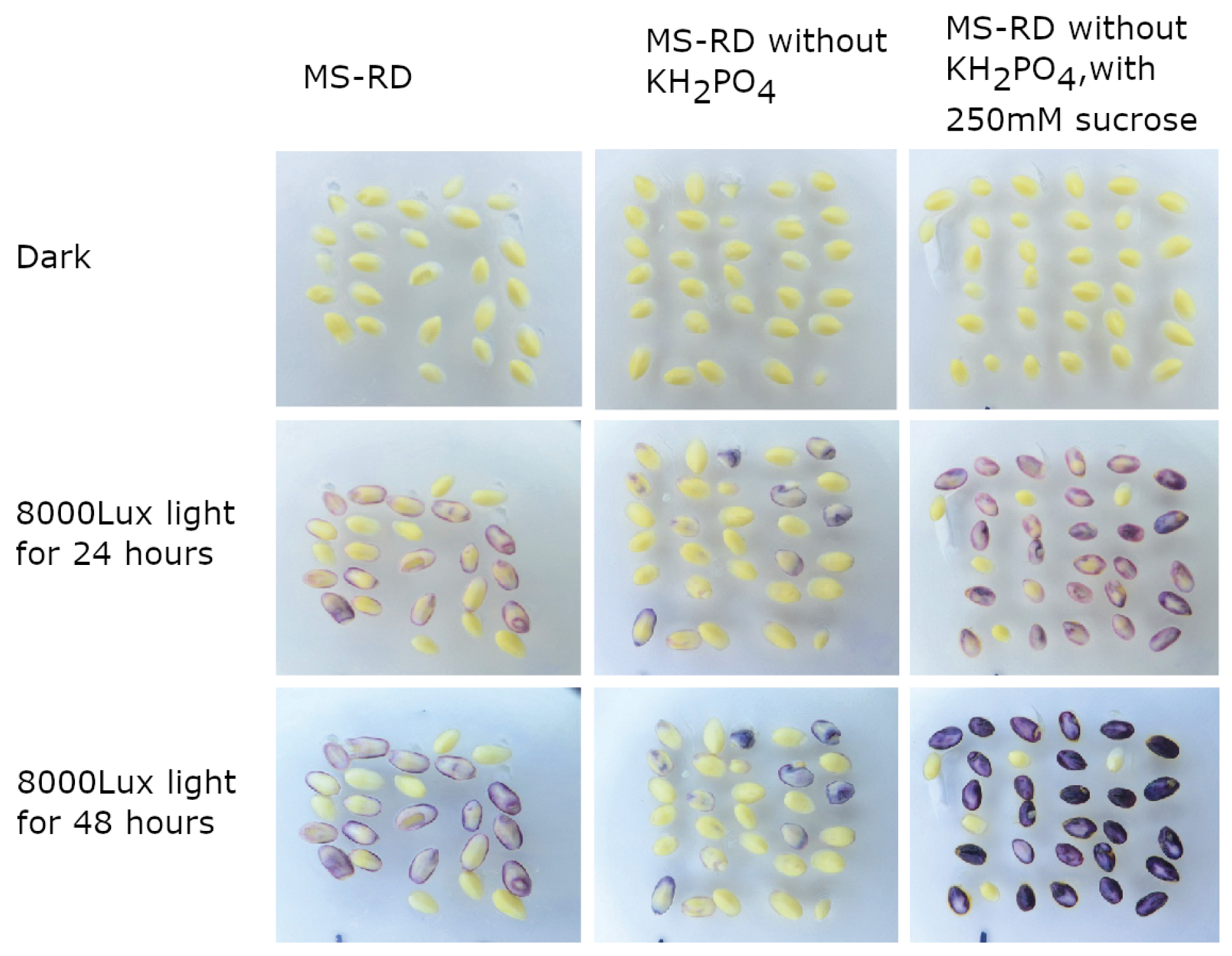
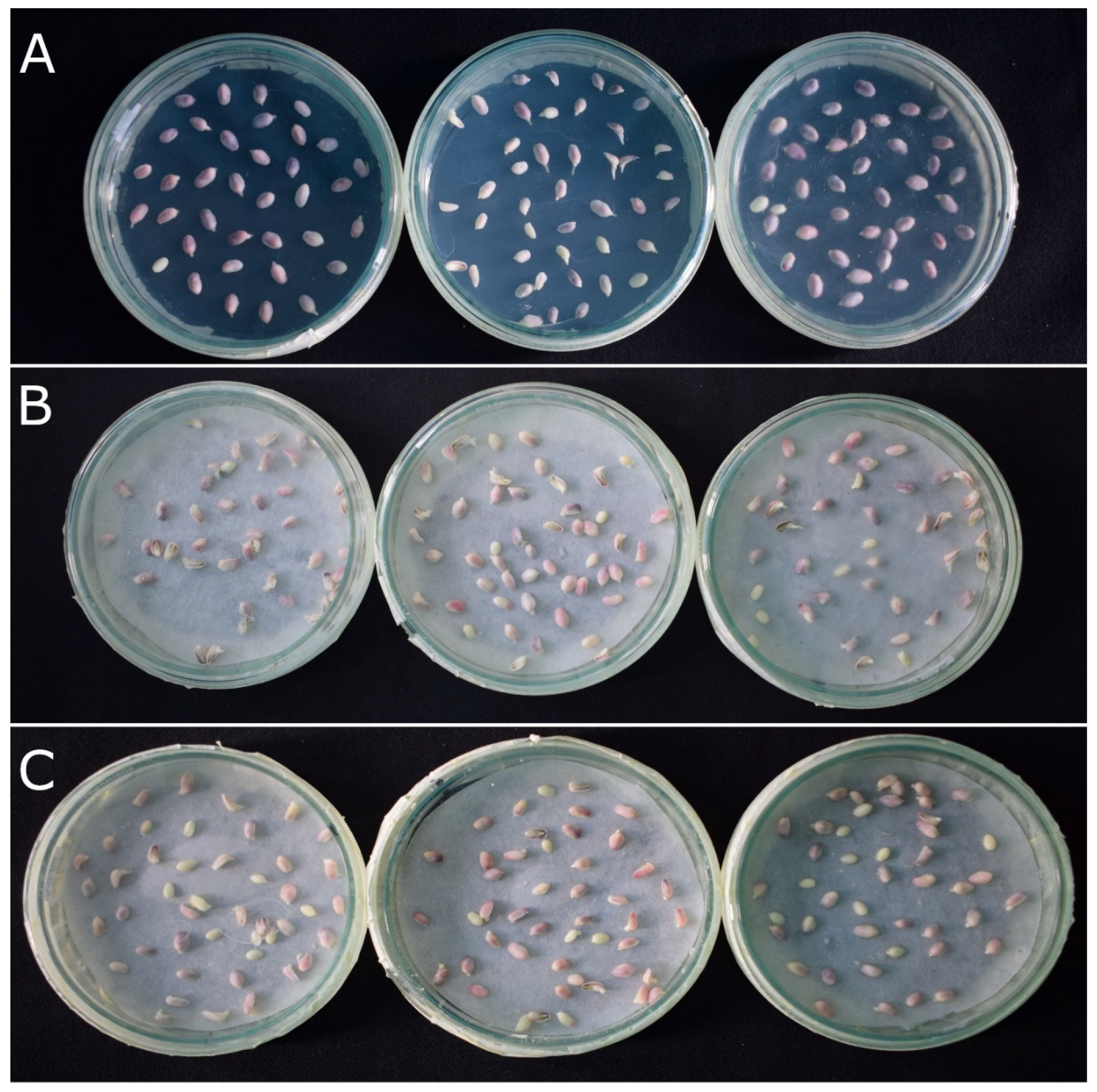
2.1. R1-nj Anthocyanin Marker Can Aid in Distinguishing Haploid Embryos from the Diploid
2.2. Verification of Accuracy of Haploid Embryo Identification Using R1-nj Marker
3. Discussion
4. Materials and Methods
4.1. Haploid Induction Crosses
4.2. Embryo Extraction
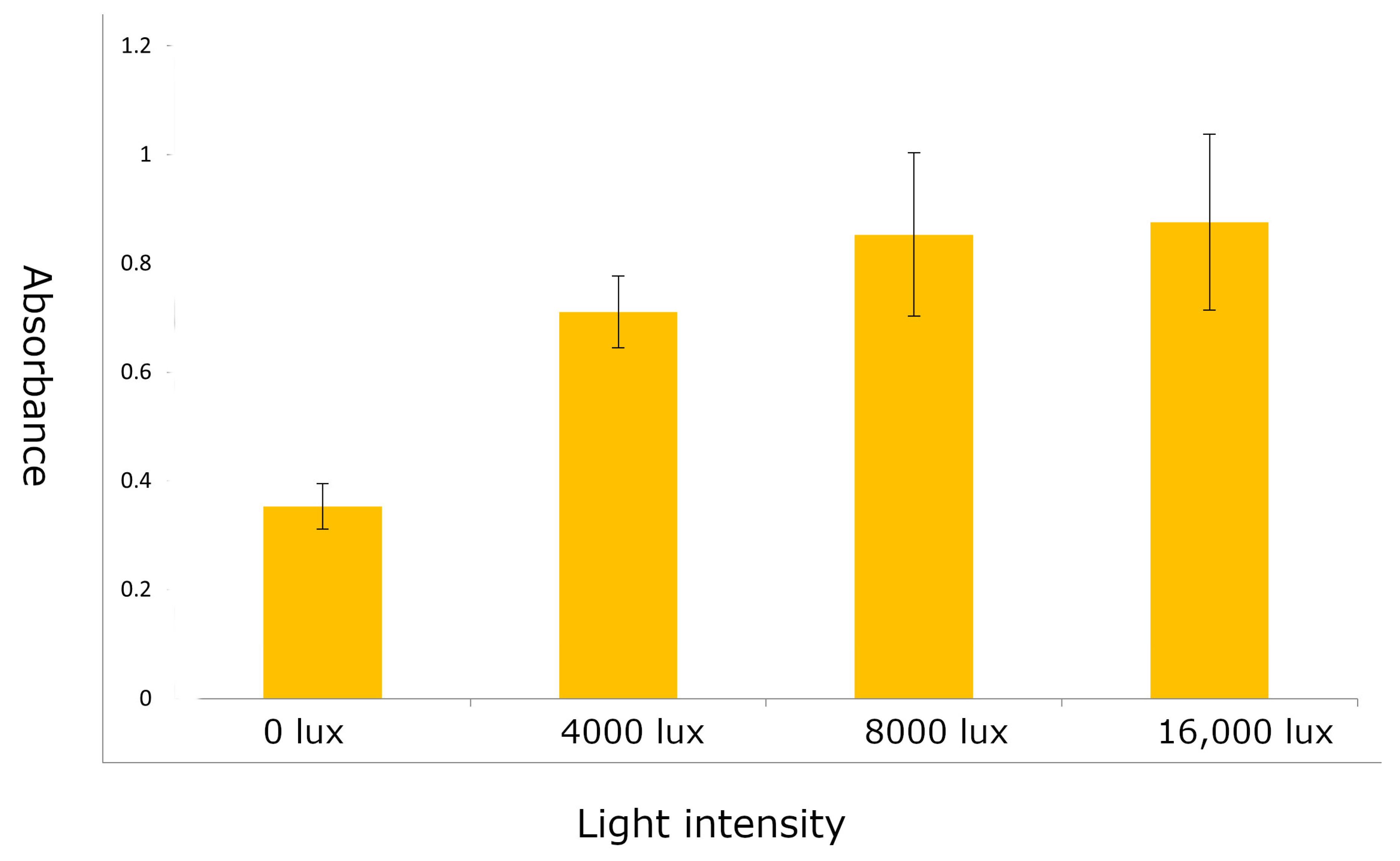
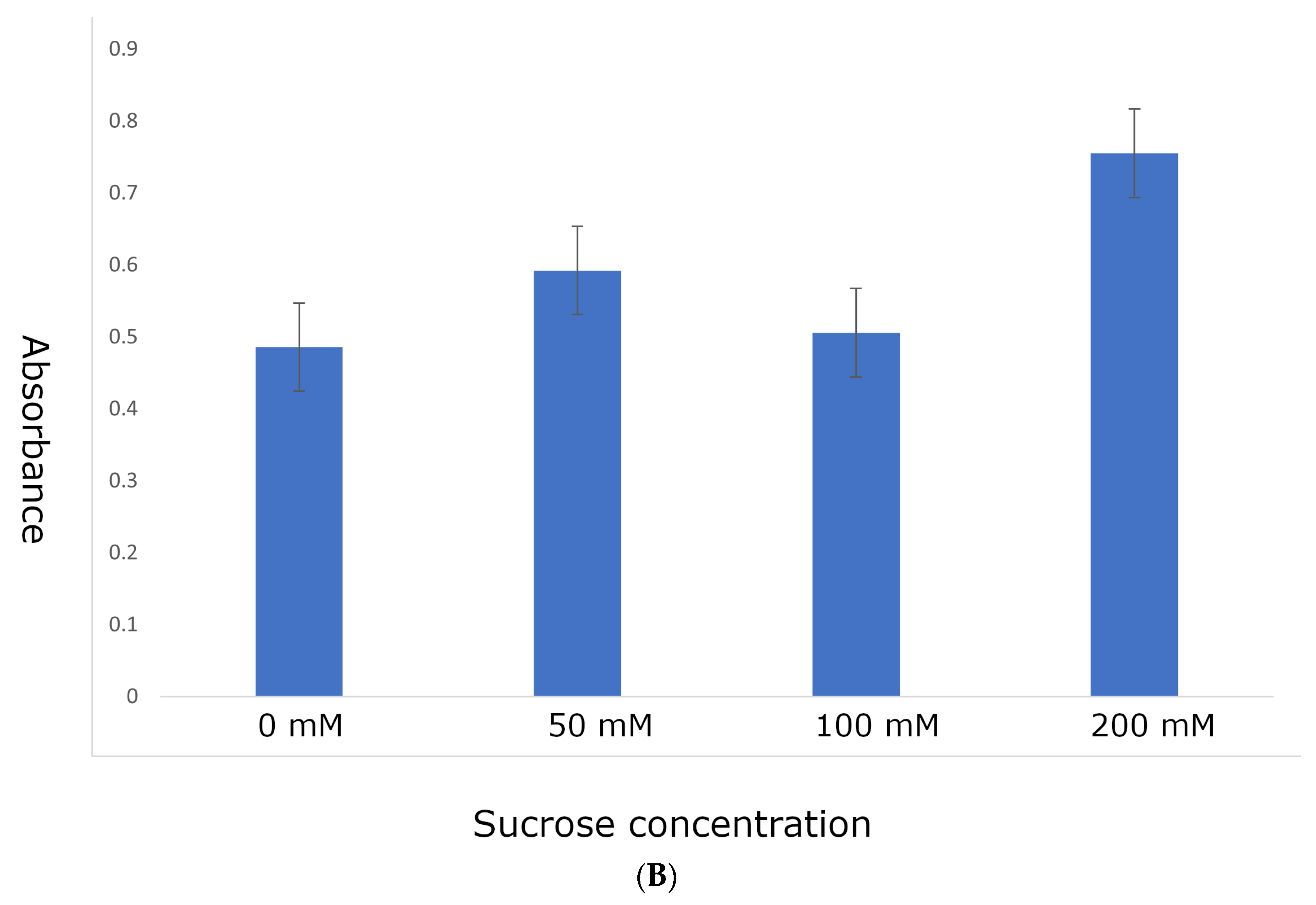
4.3. Identifying Optimal Conditions for Anthocyanin Marker Expression
4.4. Validation of R1-nj Marker-Based Haploid/Diploid Embryo Identification Was Based on a Gold Standard and Molecular Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-art and novel developments of in vivo haploid technologies. Theor. Appl. Genet. 2019, 132, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, B.M. Doubled Haploid technology in maize breeding:an overview. In Doubled Haploid Technology in Maize Breeding: Theory and Practice; Prasanna, B.M., Chaikam, V., Mahuku, G., Eds.; CIMMYT: El Batan, Mexico, 2012; pp. 1–8. [Google Scholar]
- Röber, F.K.; Gordillo, G.A.; Geiger, H.H. In vivo haploid induction in maize-performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 2005, 50, 275–283. [Google Scholar]
- Prasanna, B.M.; Chaikam, V.; Mahuku, G. Doubled Haploid Technology in Maize Breeding: Theory and Practice; CIMMYT: El Batan, Mexico, 2012; ISBN 6078263005. [Google Scholar]
- Prigge, V.; Melchinger, A.E. Production of haploids and doubled haploids in maize. In Plant Cell Culture Protocols; Loyola-Vargas, V.M., Ochoa-Alejo, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 161–172. ISBN 1617798177. [Google Scholar]
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Boddupalli, P.M. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef] [PubMed]
- Chaikam, V.; Nair, S.; Martinez, L.; Lopez, L.; Utz, H.F.; Melchinger, A.; Boddupalli, P. Marker-assisted breeding of improved maternal haploid inducers in maize for the tropical/subtropical regions. Front. Plant Sci. 2018, 9, 1527. [Google Scholar] [CrossRef] [PubMed]
- Chaikam, V.; Nair, S.K.; Babu, R.; Martinez, L.; Tejomurtula, J.; Boddupalli, P.M. Analysis of effectiveness of R1-nj anthocyanin marker for in vivo haploid identification in maize and molecular markers for predicting the inhibition of R1-nj expression. Theor. Appl. Genet. 2015, 128, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Chaikam, V.; Martinez, L.; Melchinger, A.E.; Schipprack, W.; Boddupalli, P.M. Development and validation of red root marker-based haploid inducers in maize. Crop Sci. 2016, 56, 1678–1688. [Google Scholar] [CrossRef]
- Melchinger, A.E.; Schipprack, W.; Würschum, T.; Chen, S.; Technow, F. Rapid and accurate identification of in vivo-induced haploid seeds based on oil content in maize. Sci. Rep. 2013, 3, 2129. [Google Scholar] [CrossRef]
- Melchinger, A.; Molenaar, W.; Mirdita, V.; Schipprack, W. Colchicine alternatives for chromosome doubling in maize haploids for doubled haploid production. Crop Sci. 2016, 56, 559–569. [Google Scholar] [CrossRef]
- Molenaar, W.S.; Schipprack, W.; Melchinger, A.E. Nitrous oxide-induced chromosome doubling of maize haploids. Crop Sci. 2018, 58, 650–659. [Google Scholar] [CrossRef]
- Chaikam, V.; Gowda, M.; Martinez, L.; Ochieng, J.; Omar, H.A.; Prasanna, B.M. Improving the efficiency of colchicine-based chromosomal doubling of maize haploids. Plants 2020, 9, 459. [Google Scholar] [CrossRef]
- Mongeau, D. Cultural Techniques to Increase Survivability and Vigor in Zea mays L. Doubled Haploids. Master’s Thesis, University of Delaware, Newark, DE, USA, 2010. [Google Scholar]
- Gayen, P.; Madan, J.K.; Kumar, R.; Sarkar, K.R. Chromosome doubling in haploids through colchicine. Maize Genet. Coop. Newsl. 1994, 68, 65. [Google Scholar]
- Barton, J.E.; Maddock, S.E.; Wu, X.E.; Zhao, Z.-Y.; Williams, M.E.; Hussain, T.; Gordon-Kamm, W.J. Doubling of Chromosomes in Haploid Embryos. U.S. Patent Application No. 11/532,921, 14 October 2014. [Google Scholar]
- Zhao, Z.-Y.; Bidney, D.; Elsing, E.; Miller, M.; Wu, X.; Gordon-Kamm, W. Doubled Haploid Cells, Embryos and Plants. U.S. Patent Application No. 11/298,97, 17 August 2006. [Google Scholar]
- Yu, W.; Birchler, J.A. A green fluorescent protein-engineered haploid inducer line facilitates haploid mutant screens and doubled haploid breeding in maize. Mol. Breed. 2016, 36, 5. [Google Scholar] [CrossRef]
- Dong, L.; Li, L.; Liu, C.; Liu, C.; Geng, S.; Li, X.; Huang, C.; Mao, L.; Chen, S.; Xie, C. Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Mol. Plant 2018, 11, 1214–1217. [Google Scholar] [CrossRef]
- Wang, D.; Zhong, Y.; Feng, B.; Qi, X.; Yan, T.; Liu, J.; Guo, S.; Wang, Y.; Liu, Z.; Cheng, D. The RUBY reporter enables efficient haploid identification in maize and tomato. Plant Biotechnol. J. 2023. [Google Scholar] [CrossRef]
- Chen, C.; Liu, X.; Li, S.; Liu, C.; Zhang, Y.; Luo, L.; Miao, L.; Yang, W.; Xiao, Z.; Zhong, Y. Co-expression of transcription factors ZmC1 and ZmR2 establishes an efficient and accurate haploid embryo identification system in maize. Plant J. 2022, 111, 1296–1307. [Google Scholar] [CrossRef]
- Atlin, G.N.; Cairns, J.E.; Das, B. Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob. Food Sec. 2017, 12, 31–37. [Google Scholar] [CrossRef]
- Styles, E.D.; Ceska, O.; Seah, K.-T. Developmental differences in action of R and B alleles in maize. Can. J. Genet. Cytol. 1973, 15, 59–72. [Google Scholar] [CrossRef]
- Mancinelli, A.L.; Tai, P.K.; Susinno, R. Photocontrol of anthocyanin synthesis: Phytochrome, chlorophyll and anthocyanin synthesis. Photochem. Photobiol. 1974, 20, 71–79. [Google Scholar] [CrossRef]
- Mancinelli, A.L. Light-dependent anthocyanin synthesis: A model system for the study of plant photomorphogenesis. Bot. Rev. 1985, 51, 107–157. [Google Scholar] [CrossRef]
- Mancinelli, A.L. The photoregulation of anthocyanin synthesis. In Photomorphogenesis; Springer: Berlin/Heidelberg, Germany, 1983; pp. 640–661. [Google Scholar]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Procissi, A.; Dolfini, S.; Ronchi, A.; Tonelli, C. Light-dependent spatial and temporal expression of pigment regulatory genes in developing maize seeds. Plant Cell 1997, 9, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O.; Naylor, A.W. Light control of anthocyanin biosynthesis in Zea seedlings [maize]. Physiol. Plant. 1976, 37, 62–68. [Google Scholar] [CrossRef]
- Taylor, L.P.; Briggs, W.R. Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings. Plant Cell 1990, 2, 115–127. [Google Scholar] [PubMed]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef]
- Kim, J.-S.; Lee, B.-H.; Kim, S.-H.; Oh, K.-H.; Cho, K.Y. Responses to environmental and chemical signals for anthocyanin biosynthesis in non-chlorophyllous corn (Zea mays L.) leaf. J. Plant Biol. 2006, 49, 16–25. [Google Scholar] [CrossRef]
- Leon-Cisneros, S.; Quirola-Garcés, A.; Alvarez-Santana, J.; Barriga-Medina, N.; Ramirez-Villacís, D.; Caviedes, M.; Ramirez-Cárdenas, L.; Leon-Reyes, A. Evaluation of Anthocyanin Production in White and Purple Maize (Zea mays L.) Using Methyl Jasmonate, Phosphorus Deficiency and High Concentration of Sucrose. Cereal Res. Commun. 2019, 47, 604–614. [Google Scholar] [CrossRef]
- Henry, A.; Chopra, S.; Clark, D.G.; Lynch, J.P. Responses to low phosphorus in high and low foliar anthocyanin coleus (Solenostemon scutellarioides) and maize (Zea mays). Funct. Plant Biol. 2012, 39, 255–265. [Google Scholar] [CrossRef]
- Chaikam, V.; Lopez, L.A.; Martinez, L.; Burgueño, J.; Boddupalli, P.M. Identification of in vivo induced maternal haploids in maize using seedling traits. Euphytica 2017, 213, 177. [Google Scholar] [CrossRef]
- Prigge, V.; Sánchez, C.; Dhillon, B.S.; Schipprack, W.; Araus, J.L.; Bänziger, M.; Melchinger, A.E. Doubled haploids in tropical maize: I. Effects of inducers and source germplasm on in vivo haploid induction rates. Crop Sci. 2011, 51, 1498–1506. [Google Scholar] [CrossRef]
- Rotarenco, V.A.; Dicu, G.; State, D.; Fuia, S. New inducers of maternal haploids in maize. Maize Genet. Coop News Lett. 2010, 84, 15. [Google Scholar]
- Chaikam, V.; Mahuku, G.; Prasanna, B.M. Design and implementation of maternal haploid induction. In Doubled Haploid Technoogy in Maize Breeding: Theory and Practice; Prasanna, B.M., Chaikam, V., Mahuku, G., Eds.; CIMMYT: El Batan, Mexico, 2012; pp. 14–19. [Google Scholar]
- Dilkes, B.P.; Dante, R.A.; Coelho, C.; Larkins, B.A. Genetic analyses of endoreduplication in Zea mays endosperm: Evidence of sporophytic and zygotic maternal control. Genetics 2002, 160, 1163–1177. [Google Scholar] [CrossRef]
- Gowda, M.; Worku, M.; Nair, S.K.; Palacios-Rojas, N.; Huestis, G.M.; Prasanna, B.M. Quality Assurance/Quality Control (QA/QC) in Maize Breeding and Seed Production: Theory and Practice; CIMMYT: El Batan, Mexico, 2017; ISBN 9789966197191. [Google Scholar]
| Population | Gold Standard Classification | Molecular Marker Evaluation | ||||
|---|---|---|---|---|---|---|
| N | FDR | FNR | N | FDR | FNR | |
| (CKDHL0262/LH132)-B | 276 | 30.0 | 0.0 | 30 | 0.0 | 0.0 |
| (CKLMARS1C3S50264/PHW52)-B | 180 | 43.4 | 3.2 | 32 | 75.0 | 20.0 |
| (CKDHL0378/PHR03)-B | 171 | 8.3 | 0.0 | 32 | 0.0 | 0.0 |
| (CKDHL120390/LH198)-B | 208 | 17.1 | 0.0 | 32 | 0.0 | 0.0 |
| CKIR04003 | 144 | 28.6 | 6.3 | 32 | 25.0 | 7.7 |
| CKDHL164288/CKDHL166062 | 203 | 12.3 | 3.0 | 32 | 0.0 | 11.1 |
| 1182 | 23.5 | 1.2 | 190 | 16.8 | 2.0 | |
| Population | N | FDR |
|---|---|---|
| CZDHL150813/CKDHL164029 | 611 | 8.2 |
| CZDHL150813/CKDHL163669 | 437 | 13.3 |
| CZDHL151801/CKDHL163629 | 302 | 11.6 |
| CKDHL165841/CKDHL166103 | 289 | 5.9 |
| CZDHL151890/CKDHL163629 | 270 | 17.8 |
| CKDHL150363/CKDHL152036 | 242 | 7.4 |
| CZDHL153829/CKDHL163629 | 191 | 23.0 |
| CKDHL0165/KS23-6 | 180 | 12.2 |
| (CKL147/N21)-B | 140 | 30.0 |
| CZDHL155303/CZDHL154584 | 139 | 10.8 |
| (CKL15117/PHAP1)-B | 131 | 19.8 |
| ((CML536/CML463)DH173-B-B-1-B-B/PHW61)-B | 128 | 35.9 |
| (CKL15117/PHAP1)-B | 82 | 4.9 |
| (CKL1574/CL114162)-B | 82 | 17.1 |
| CZDHL153829/CKDHL164029 | 79 | 20.3 |
| (DJL173833/PHAP1)-B | 76 | 11.8 |
| TZMI717/CML498 | 73 | 12.3 |
| CKDHL153508/CKDHL164461 | 72 | 8.3 |
| CML540LNT22/CKDHL0186LNT23 | 56 | 82.1 |
| ((CL114162/KS23-6)/CL114162)-B | 53 | 43.4 |
| (CKL141999/CL1211293)-B | 48 | 8.3 |
| CML572LNT23/CKL05017LNT22 | 40 | 20.0 |
| ((CML536/CML463)DH173-B-B-1-B-B/PHTV7)-B | 33 | 12.1 |
| TZMI717/NMCOMPOSITE2008-4-2-1-1-2-1 | 21 | 28.6 |
| Average FDR | 19.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez, L.A.; Ochieng, J.; Pacheco, M.; Martinez, L.; Omar, H.A.; Gowda, M.; Prasanna, B.M.; Dhugga, K.S.; Chaikam, V. Effectiveness of R1-nj Anthocyanin Marker in the Identification of In Vivo Induced Maize Haploid Embryos. Plants 2023, 12, 2314. https://doi.org/10.3390/plants12122314
Lopez LA, Ochieng J, Pacheco M, Martinez L, Omar HA, Gowda M, Prasanna BM, Dhugga KS, Chaikam V. Effectiveness of R1-nj Anthocyanin Marker in the Identification of In Vivo Induced Maize Haploid Embryos. Plants. 2023; 12(12):2314. https://doi.org/10.3390/plants12122314
Chicago/Turabian StyleLopez, Luis Antonio, John Ochieng, Mario Pacheco, Leocadio Martinez, Hamilton Amoshe Omar, Manje Gowda, Boddupalli M. Prasanna, Kanwarpal S. Dhugga, and Vijay Chaikam. 2023. "Effectiveness of R1-nj Anthocyanin Marker in the Identification of In Vivo Induced Maize Haploid Embryos" Plants 12, no. 12: 2314. https://doi.org/10.3390/plants12122314
APA StyleLopez, L. A., Ochieng, J., Pacheco, M., Martinez, L., Omar, H. A., Gowda, M., Prasanna, B. M., Dhugga, K. S., & Chaikam, V. (2023). Effectiveness of R1-nj Anthocyanin Marker in the Identification of In Vivo Induced Maize Haploid Embryos. Plants, 12(12), 2314. https://doi.org/10.3390/plants12122314






