Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii
Abstract
1. Introduction
2. Results
2.1. Flow Cytometry
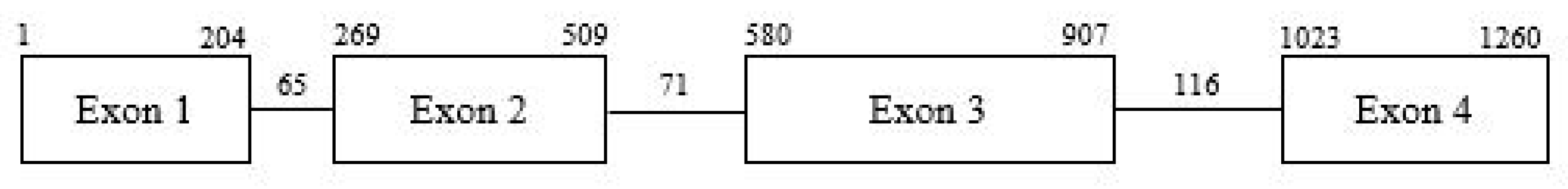
2.2. Identification of ObDMR6 in ‘FT Italiko’ and Protein Alignment
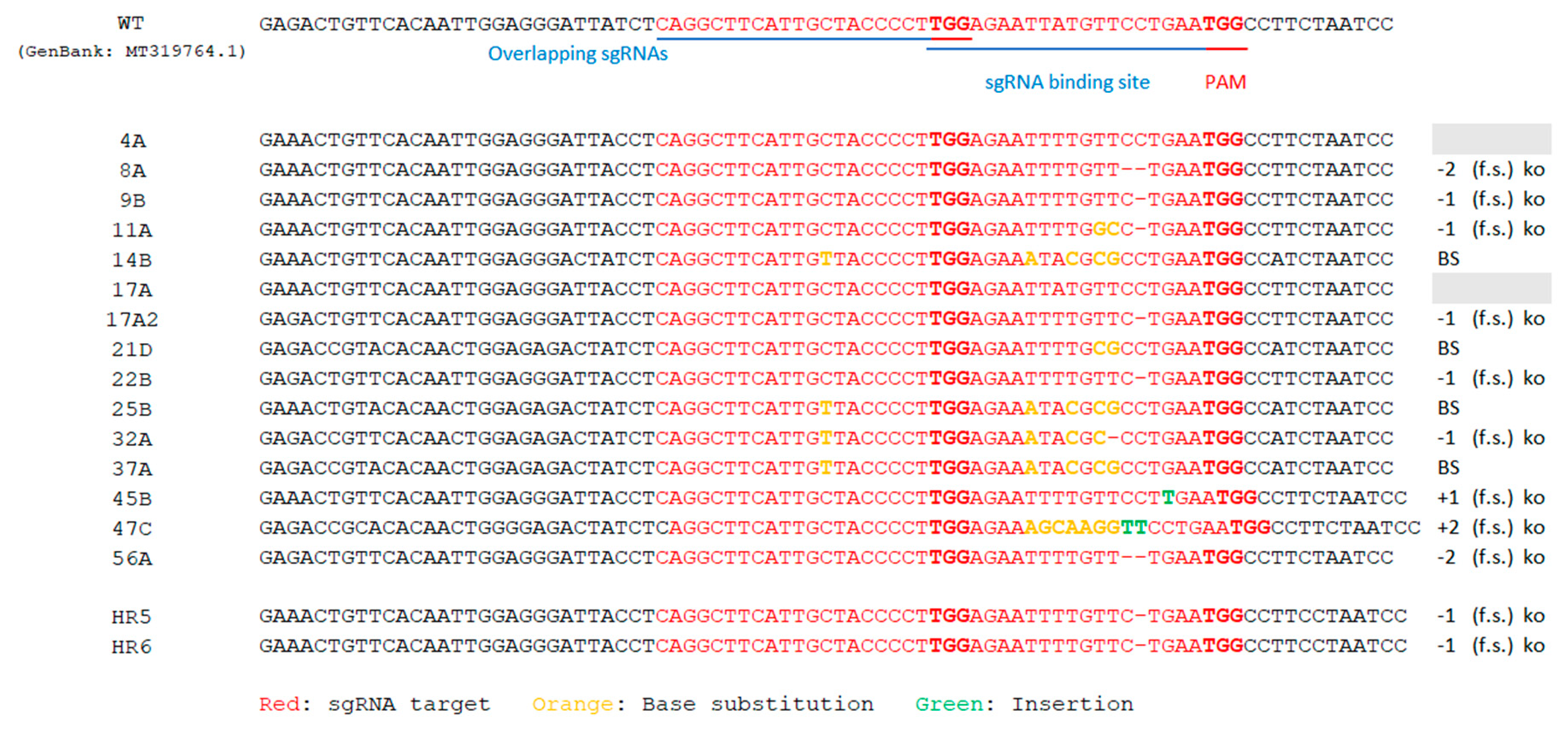
2.3. Editing in Hairy Roots
2.4. Production of T0 Edited Plants by Agrobacterium Tumefaciens-Mediated Transformation
2.5. Cas9-Free ObDMR6 Mutants in T1 Generation
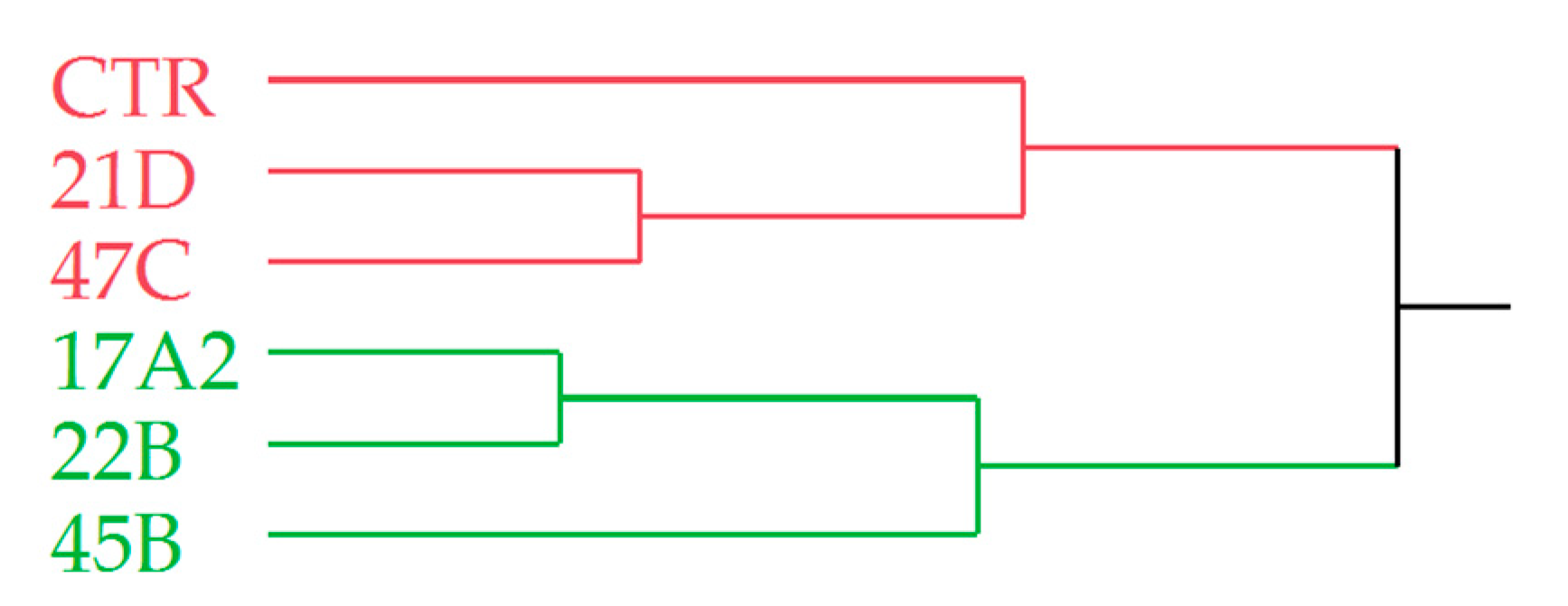
2.6. Morphological Phenotyping of T0 and T1 Plants
2.7. P. belbahrii Infection Trial
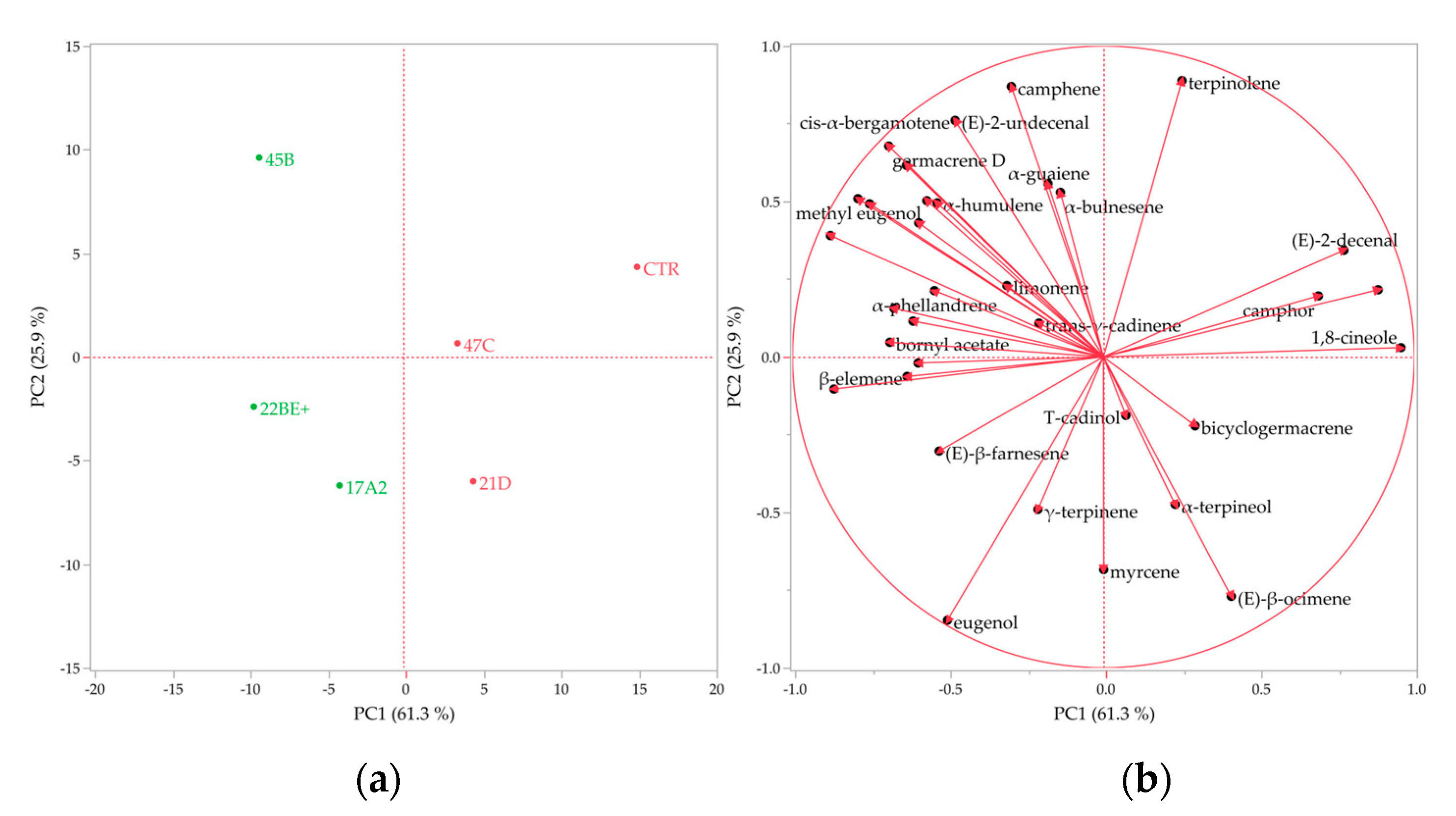
2.8. Volatile Profile Analysis
3. Discussion
4. Materials and Methods
4.1. Flow Cytometry
4.2. Regeneration Protocol Via Direct Organogenesis and Agrobacterium Tumefaciens Mediated Transformation
4.3. Agrobacterium Rhizogenes Mediated Transformation
4.4. Identification of ObDMR6 in ‘FT Italiko’
4.5. Protein Alignment
4.6. Identification of CRISPR/Cas9 Targets and Vector Construction
4.7. Screening of Transgenic Plants and HRs for Target ObDMR6 Mutations
4.8. Phenotypic Evaluation of T0 and T1 Mutant Plants
4.9. Evaluation of In Vivo T0 Plants Infection
4.10. Volatile Profile Analysis
4.10.1. Headspace Solid Phase Microextraction Analysis
4.10.2. Gas Chromatography–Mass Spectrometry Analyses
4.10.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pyne, R.M.; Honig, J.A.; Vaiciunas, J.; Wyenandt, C.A.; Simon, J.E. Population structure, genetic diversity and downy mildew resistance among Ocimum species germplasm. BMC Plant Biol. 2018, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Koroch, A.R.; Wang, W.; Dudai, N.; Belanger, F. Estimation of nuclear DNA content of cultivated Ocimum species by using flow cytometry. Isr. J. Plant Sci. 2010, 58, 183–189. [Google Scholar] [CrossRef]
- Da Costa, A.S.; Arrigoni-Blank, M.d.F.; Carvalho Filho, J.L.S.D.; de Santana, A.D.D.; Santos, D.d.A.; Alves, P.B.; Blank, A.F. Chemical diversity in basil (Ocimum sp.) germplasm. Sci. World J. 2015, 2015, 352638. [Google Scholar] [CrossRef] [PubMed]
- Wyenandt, C.A.; Simon, J.E.; Pyne, R.M.; Homa, K.; McGrath, M.T.; Zhang, S.; Raid, R.N.; Ma, L.J.; Wick, R.; Guo, L.; et al. Basil downy mildew (Peronospora belbahrii): Discoveries and challenges relative to its control. Phytopathology 2015, 105, 885–894. [Google Scholar] [CrossRef]
- Ekmekci, H.; Aasim, M. In vitro plant regeneration of Turkish sweet basil (Ocimum basilicum L.). J. Anim. Plant Sci. 2014, 24, 1758–1765. [Google Scholar]
- Li, Q.X.; Chang, C.L. Basil (Ocimum basilicum L.) oils. In Essential Oils in Food Preservation, Flavor and Safety; Preedy, V.R., Ed.; Academic Press: London, UK, 2016; pp. 231–238. [Google Scholar] [CrossRef]
- Gilardi, G.; De Marchi, S.; Garibaldi, A.; Gullino, M.L. Management of downy mildew of sweet basil (Ocimum basilicum) caused by Peronospora belbahrii by means of resistance inducers, fungicides, biocontrol agents and natural products. Phytoparasitica 2013, 41, 59–72. [Google Scholar] [CrossRef]
- Collina, M.; Merighi, M.; Turan, C.; Pirondi, A.; Minuto, G.; Brunelli, A. First report of resistance of Peronospora belbahrii, causal agent of downy midew of basil, to mefenoxam in Italy. Plant Dis. 2016, 100, 1787. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gullino, M.L.; Minuto, G. Diseases of basil and their management. Plant Dis. 1997, 81, 124–132. [Google Scholar] [CrossRef]
- Gopi, C.; Sekhar, Y.N.; Ponmurugan, P. In vitro multiplication of Ocimum gratissimum L. through direct regeneration. Afr. J. Biotechnol. 2006, 5, 723–726. [Google Scholar]
- Homa, K.; Barney, W.P.; Ward, D.L.; Wyenandt, C.A.; Simon, J.E. Morphological characteristics and susceptibility of basil species and cultivars to Peronospora belbarii. HortScience 2016, 51, 1389–1396. [Google Scholar] [CrossRef]
- Cohen, Y.; Ben Naim, Y.; Falach, L.; Rubin, A.E. Epidemiology of basil downy mildew. Phytopatology 2017, 107, 1149–1160. [Google Scholar] [CrossRef]
- Cooper, J.B. An Evaluation of Eight Basil Cultivars for Downy Mildew Resistance in Virginia. Master’s Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 2019. Available online: https://vtechworks.lib.vt.edu/bitstream/handle/10919/96224/Jason%20Cooper%20Basil%20Project%20Report.pdf?sequence=1&isAllowed=y (accessed on 15 December 2022).
- Phippen, W.B.; Simon, J.E. Shoot regeneration of young leaf explants from basil (Ocimum basilicum L.). Vitr. Cell. Dev. Biol. Plant. 2000, 36, 250–254. [Google Scholar] [CrossRef]
- Ben-Naim, Y.; Falach, L.; Cohen, Y. Transfer of downy mildew resistance from wild basil (Ocimum americanum) to sweet basil (O. basilicum). Phytopathology 2018, 108, 114–123. [Google Scholar] [CrossRef]
- Gupta, S.; Srivastava, A.; Shasany, A.K.; Gupta, A.K. Genetics, Cytogenetics, and Genetic Diversity in the Genus Ocimum. In The Ocimum Genome; Shasany, A.K., Kole, C., Eds.; Compendium of Plant Genomes; Springer International Publishing: Cham, Switzerland, 2018; pp. 73–78. ISBN 978-3-319-97429-3. [Google Scholar]
- Van Schie, C.C.N.; Takken, F.L.W. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopatholol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Smiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 617. [Google Scholar] [CrossRef]
- Pan, C.T.; Ye, L.; Qin, L.; Liu, X.; He, Y.J.; Wang, J.; Chen, L.F.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Wang, F.J.; Wang, C.L.; Liu, P.Q.; Lei, C.L.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K.J. Enhanced rice blast resistance by CRISPR/Cas9-Targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Gumtow, R.; Wu, D.L.; Uchida, J.; Tian, M.Y. A Phytophthora palmivora extracellular Cystatin-Like protease inhibitor targets papain to contribute to virulence on Papaya. Mol. Plant Microbe Interact. 2018, 31, 363–373. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H.; Szurek, B.; Van den Ackerveken, G. Stop helping pathogens: Engineering plant susceptibility genes for durable resistance. Curr. Opin. Biotech. 2021, 70, 187–195. [Google Scholar] [CrossRef]
- Van Damme, M.; Andel, A.; Huibers, R.P.; Panstruga, R.; Weisbeek, P.J.; Van den Ackerveken, G. Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol. Plant Microbe Interact 2005, 18, 583–592. [Google Scholar] [CrossRef]
- Van Damme, M.; Huibers, R.P.; Elberse, J.; Van den Ackerveken, G. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van den Ackerveken, G. Downy mildew resistant 6 and DMR6-like oxygenase 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015, 81, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Wolters, A.M.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.; Jacobsen, E.; Visser, R.G.; Bai, Y. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 2016, 25, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Pirrello, C.; Zeilmaker, T.; Bianco, L.; Giacomelli, L.; Moser, C.; Vezzulli, S. Mining grapevine downy mildew susceptibility genes: A resource for genomics-based breeding and tailored gene editing. Biomolecules 2021, 11, 181. [Google Scholar] [CrossRef]
- Pavese, V.; Moglia, A.; Gonthier, P.; Torello Marinoni, D.; Cavalet-Giorsa, E.; Botta, R. Identification of susceptibility genes in Castanea sativa and their transcription dynamics following pathogen infection. Plants 2021, 10, 913. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The identification of maize and Arabidopsis type I FLAVONE SYNTHASEs links flavones with hormones and biotic interactions. Plant Physiol. 2015, 169, 1090–1107. [Google Scholar] [CrossRef]
- Thomazella, D.P.D.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.C.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Low, Y.C.; Lawton, M.A.; Di, R. Validation of barley 2OGO gene as a functional orthologue of Arabidopsis DMR6 gene in Fusarium head blight susceptibility. Sci. Rep. 2020, 10, 993. [Google Scholar] [CrossRef]
- Tripathi, J.; Ntui, V.; Tripathi, L. Brief Communication CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotech. J. 2021, 19, 1291–1293. [Google Scholar] [CrossRef]
- Kieu, N.P.; Lenman, M.; Wang, E.S.; Petersen, B.L.; Andreasson, E. Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci Rep. 2021, 24, 4487. [Google Scholar] [CrossRef]
- Shan, D.; Wang, C.; Zheng, X.; Hu, Z.; Zhu, Y.; Zhao, Y.; Jiang, A.; Zhang, H.; Shi, K.; Bai, Y.; et al. MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 2021, 186, 1202–1219. [Google Scholar] [CrossRef]
- Parajuli, S.; Huo, H.; Gmitter, F.G.; Duan, Y.; Luo, F.; Deng, Z. Editing the CsDMR6 gene in citrus results in resistance to the bacterial disease citrus canker. Hortic Res. 2022, 11, uhac082. [Google Scholar] [CrossRef]
- Giacomelli, L.; Zeilmaket, T.; Scintilla, S.; Salvagnin, U.; van der Voort, J.R.; Moser, C. Vitis vinifera plants edited in DMR6 genes show improved resistance to downy mildew. bioRxiv 2022, 2022, 488768. [Google Scholar]
- Hasley, J.A.R.; Navet, N.; Miaoying, T. CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PLoS ONE 2021, 16, e0253245. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Teshome, D.T.; Zharare, G.E.; Naidoo, S. The Threat of the Combined Effect of Biotic and Abiotic Stress Factors in Forestry Under a Changing Climate. Front Plant Sci. 2020, 11, 1874. [Google Scholar] [CrossRef]
- Pirrello, C.; Malacarne, G.; Moretto, M.; Lenzi, L.; Perazzolli, M.; Zeilmaker, T.; Van den Ackerveken, G.; Pilati, S.; Moser, C.; Giacomelli, L. Grapevine DMR6-1 Is a Candidate Gene for Susceptibility to Downy Mildew. Biomolecules 2022, 12, 182. [Google Scholar] [CrossRef]
- Dima, O.; Heyvaert, Y.; Inzé, D. Interactive database of genome editing applications in crops and future policy making in the European Union. Trend. Plant Sci. 2022, 27, 746–748. [Google Scholar] [CrossRef]
- Hsieh, Y.F.; Jain, M.; Wang, J.; Gallo, M. Direct organogenesis from cotyledonary node explants suitable for Agrobacterium-mediated transformation in peanut (Arachis hypogaea L.). Plant Cell Tissue Organ Cult. 2017, 128, 161–175. [Google Scholar] [CrossRef]
- Abahmane, L. Cultivar-Dependent Direct Organogenesis of Date Palm from Shoot Tip Explants. Methods Mol. Biol. 2017, 1637, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Bahramnejad, B.; Naji, M.; Bose, R.; Jha, S. A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnol. Adv. 2019, 37, 107405. [Google Scholar] [CrossRef] [PubMed]
- Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants 2022, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; López, C.M.; Martínez Rivas, F.J.; Torralbo, F.; Bulut, M.; Alseekh, S. Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein and hairy roots: A perfect match for gene functional analysis and crop improvement. Curr. Opin. Biotechnol. 2023, 79, 102876. [Google Scholar] [CrossRef]
- Gonda, I.; Faigenboim, A.; Adler, C.; Milavski, R.; Karp, M.J.; Shachter, A.; Ronen, G.; Baruch, K.; Chaimovitsh, D.; Dudai, N. The genome sequence of tetraploid sweet basil, Ocimum basilicum L., provides tools for advanced genome editing and molecular breeding. DNA Res. 2020, 27, dsaa027. [Google Scholar] [CrossRef]
- Carović-Stanko, K.; Liber, Z.; Politeo, O.; Strikić, F.; Kolak, I.; Milos, M.; Satovic, Z. Molecular and chemical characterization of the most widespread Ocimum species. Plant Syst. Evol. 2011, 294, 253–262. [Google Scholar] [CrossRef]
- Jang, D.E.; Lee, J.Y.; Lee, J.H. Multiple sgRNAs with overlapping sequences enhance CRISPR/Cas9-mediated knock-in efficiency. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef]
- Navet, N.; Tian, M.Y. Efficient targeted mutagenesis in allotetraploid sweet basil by CRISPR/Cas9. Plant Direct 2020, 4, e00233. [Google Scholar] [CrossRef]
- Zhang, X.; Low, Y.C.; Lawton, M.A.; Simon, J.E.; Di, R. CRISPR-Editing of Sweet Basil (Ocimum basilicum L.) Homoserine Kinase Gene for Improved Downy Mildew Disease Resistance. Front. Genome Ed. 2021, 3, 629769. [Google Scholar] [CrossRef]
- Tarchoune, I.; Baâtour, O.; Harrathi, J.; Cioni, P.L.; Lachaâl, M.; Flamini, G.; Ouerghi, Z. Essential oil and volatile emissions of basil (Ocimum basilicum) leaves exposed to NaCl or Na2SO4 salinity. Z. Pflanzenernähr. Bodenk. 2013, 176, 748–755. [Google Scholar] [CrossRef]
- Monfort, L.E.F.; Bertolucci, S.K.V.; Lima, A.F.; de Carvalho, A.A.; Mohammed, A.; Blank, A.F.; Pinto, J.E.B.P. Effects of plant growth regulators, different culture media and strength MS on production of volatile fraction composition in shoot cultures of Ocimum basilicum. Ind. Crops Prod. 2018, 116, 231–239. [Google Scholar] [CrossRef]
- Keng Hong, T.; Ritsuo, N. Methyl eugenol: Its occurrence, distribution, and role in nature, especially in relation to insect behavior and pollination. J. Insect Sci. 2012, 12, 56. [Google Scholar] [CrossRef]
- Oltmanns, H.; Frame, B.; Lee, L.Y.; Johnson, S.; Li, B.; Wang, K.; Gelvin, S.B. Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 2010, 152, 1158–1166. [Google Scholar] [CrossRef]
- Johnson, E.T.; Kim, H.S.; Tian, M.; Dudai, N.; Ta, O.l.; Gonda, I. Dual transcriptional analysis of Ocimum basilicum and Peronospora belbahrii in susceptible interactions. Plant Gene 2022, 29, 100350. [Google Scholar] [CrossRef]
- Omidbaigi, R.; Mirzaee, M.; Hassani, M.; Sedghi Moghadam, M. Induction and identification of polyploidy in basil (Ocimum basilicum L.) medicinal plant by colchicine treatment. Int. J. Plant Prod. 2012, 4, 87–98. [Google Scholar] [CrossRef]
- White, P.; Buckner, E.; Pitilla, J.; Cogbill, C. High-elevation forests: Spruce-fir forests, northern hardwood forests, and associated communities. In Biodiversity of the Southeastern United States: Upland Terrestrial Communities; Martin, W., Boyce, S., Echternacht, A., Eds.; Wiley: New York, NY, USA, 1993; pp. 305–338. [Google Scholar]
- Forti, C.; Barberini, S.; Laura, M.; Ciorba, R.; Mascarello, C.; Giovannini, A.; Ruffoni, B.; Savona, M. Messa a punto di protocolli di rigenerazione in vitro in Ocimum basilicum cv FT Italiko, finalizzati al miglioramento genetico via genome editing. Atti Giornate Sci. SOI Bari 2022. submitted. [Google Scholar]
- Barberini, S.; Forti, C.; Laura, M.; Ciorba, R.; Mascarello, C.; Giovannini, A.; Ruffoni, B.; Savona, M. An Optimized Protocol for In Vitro Regeneration of Ocimum basilicum cv. FT Italiko. Horticulturae 2023, 9, 407. [Google Scholar] [CrossRef]
- Rastogi, S.; Shah, S.; Kumar, R.; Vashisth, D.; Akhtar, M.Q.; Kumar, R.; Vashisth, D.; Md Akhtar, M.Q.; Kumar, A.; Dwivedi, U.N.; et al. Ocimum metabolomics in response to abiotic stresses: Cold, flood, drought, and salinity. PLoS ONE 2019, 14, e0210903. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; Voytas, D.F. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [PubMed]
- Lazo, G.R.; Stein, P.A.; Ludwig, R.A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 1991, 9, 963–967. [Google Scholar] [CrossRef] [PubMed]
- An, G. Binary ti-vectors for plant transformation and promoter analysis. Methods Enzymol. 1987, 153, 292–305. [Google Scholar]
- Adams, R.P.R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectroscopy; Carol, S., Ed.; Allured Publishing Corporation: Carol Stream, IL, USA, 2007; ISBN 1932633219. [Google Scholar]




| Leaf Traits | Plant Traits | |||||
|---|---|---|---|---|---|---|
| Clones | Morphology | Size (cm) 3rd True Leaf | Height (cm) | Internode Number | Internode Length (cm) | Flowering Attitude |
| WT | Ovate with moderately serrated margin | Length: 7.75 ± 0.35 Width: 4.25 ± 0.35 | 48.5 ± 4.95 | 12.0 ± 0.41 | 4.04 ± 0.41 | Yes |
| 22B | Few leaves with curling up margin | Similar to WT | Similar to WT | Similar to WT | Similar to WT | Yes |
| 21D | Few leaves with curling up margin | Shorter with decreased length/width ratio | Strongly reduced | Decreased | Shorter | Yes |
| 45B | Similar to WT | Shorter with decreased length/width ratio | Shorter | Decreased | Similar to WT | Yes |
| 17A2 | Few leaves with curling up margin | Similar to WT | Shorter | Decreased | Similar to WT | No |
| 47C | Few leaves with curling up margin | Shorter with decreased length/width ratio | Strongly reduced | Decreased | Shorter | No |
| 32A | Few leaves with curling up margin | Similar to WT | Shorter | Similar to WT | Shorter | No |
| Leaf Traits | Plant Traits | ||||
|---|---|---|---|---|---|
| Progenies | Morphology | Size (cm) 3rd True Leaf | Height (cm) | Internode Number | Internode Length (cm) |
| WT | Ovate with moderately serrated margin | Length: 6.4 ± 0.89 Width: 4.5 ± 0.35 | 27.60 ± 3.29 | 7.40 ± 0.55 | 3.73 ± 0.26 |
| 22B | Similar to WT | Higher | Similar to WT | Similar to WT | Longer |
| 21D | Few leaves with curling up margin | Similar to WT | Similar to WT | Similar to WT | Similar to WT |
| Clone | 7 dpi | 10 dpi | 14 dpi |
|---|---|---|---|
| WT | 10.0 ab | 96.7 e | 100.0 b |
| 17A2 | 10.0 ab | 93.3 e | 100.0 b |
| 47C | 50.0 d | 75.0 d | 100.0 b |
| 21D | 23.1 c | 48.7 c | 100.0 b |
| 45B | 20.0 bc | 23.3 b | 50.0 a |
| 22B | 0.0 a | 6.7 a | 41.7 a |
| Clone | 7 dpi | 10 dpi | 14 dpi |
|---|---|---|---|
| WT | 28.3 bc | 90.0 d | 100.0 b |
| 17A2 | 43.3 c | 90.0 d | 100.0 b |
| 47C | 30.0 bc | 60.0 c | 100.0 b |
| 21D | 33.3 bc | 65.0 c | 100.0 b |
| 45B | 20.0 bc | 21.7 b | 45.0 a |
| 22B | 0.0 a | 10.0 a | 43.3 a |
| Compounds | l.r.i. 1 | Class. | Relative Abundance ± Standard Deviation (n = 3) | |||||
| WT/CTR | 17A2 | 21D | 22B | 45B | 47C | |||
| α-pinene | 933 | mh | 0.8 ± 0.01 | 1.5 ± 0.05 | 1.6 ± 0.25 | 1.9 ± 0.27 | 2.2 ± 0.21 | 2.3 ± 0.78 |
| camphene | 948 | mh | 0.5 ± 0.03 | 0.4 ± 0.03 | 0.3 ± 0.02 | 0.5 ± 0.05 | 0.7 ± 0.01 | 0.6 ± 0.21 |
| sabinene | 973 | mh | 1.3 ± 0.05 | 1.9 ± 0.11 | 2., ± 0.4 | 2.0 ± 0.25 | 2.2 ± 0.26 | 2.4 ± 0.55 |
| β-pinene | 977 | mh | 2.2 ± 0.06 | 3.5 ± 0.17 | 3.6 ± 0.72 | 3.9 ± 0.43 | 4.5 ± 0.54 | 4.9 ± 1.40 |
| myrcene | 991 | mh | 1.7 ± 0.02 | 2.8 ± 0.15 | 3.1 ± 0.25 | 2.7 ± 0.00 | 1.6 ± 0.57 | 3.7 ± 0.98 |
| α-phellandrene | 1006 | mh | - 2 | 0.1 ± 0.02 | - | - | 0.1 ± 0.02 | - |
| δ-3-carene | 1011 | mh | - | 0.1 ± 0.01 | - | - | 0.2 ± 0.03 | - |
| limonene | 1029 | mh | 1.4 ± 0.01 | 1.5 ± 0.2 | 1.6 ± 0.43 | 1.7 ± 0.18 | 1.8 ± 0.03 | 2.1 ± 0.31 |
| 1,8-cineole | 1031 | om | 44.1 ± 0.4 | 34.2 ± 6.02 | 41.2 ± 8.3 | 29.9 ± 1.79 | 33.1 ± 5.92 | 41.0 ± 4.21 |
| (E)-β-ocimene | 1047 | mh | 7.3 ± 0.00 | 7.9 ± 1.10 | 11.2 ± 1.33 | 6.3 ± 0.70 | 3.2 ± 1.19 | 4.8 ± 0.97 |
| γ-terpinene | 1058 | mh | - | 0.2 ± 0.06 | - | - | - | - |
| terpinolene | 1089 | mh | 1.5 ± 0.01 | 1.2 ± 0.14 | 1.2 ± 0.36 | 1.3 ± 0.05 | 1.5 ± 0.06 | 1.5 ± 0.21 |
| linalool | 1101 | om | 16.6 ± 0.26 | 2.9 ± 0.06 | 3.9 ± 1.47 | 1.8 ± 1.34 | 1.0 ± 0.56 | 4.2 ± 0.65 |
| camphor | 1145 | om | 3.5 ± 0.02 | 1.8 ± 0.22 | 1.7 ± 0.19 | 1.7 ± 0.33 | 1.5 ± 0.61 | 1.2 ± 0.54 |
| α-terpineol | 1191 | om | 0.3 ± 0.03 | 0.3 ± 0.03 | 0.4 ± 0.04 | 0.3 ± 0.09 | 0.3 ± 0.03 | 0.3 ± 0.04 |
| (E)-2-decenal | 1261 | nt | 0.1 ± 0.07 | - | - | - | - | - |
| bornyl acetate | 1286 | om | - | 0.2 ± 0.09 | 0.2 ± 0.23 | 0.2 ± 0.02 | 0.3 ± 0.32 | 0.3 ± 0.03 |
| (E)-methyl geranate | 1325 | om | - | - | - | - | 0.2 ± 0.06 | - |
| eugenol | 1357 | pp | 7.0 ± 0.02 | 20.0 ± 5.6 | 15.4 ± 7.38 | 18.7 ± 2.96 | 8.6 ± 2.41 | 11.2 ± 3.48 |
| (E)-2-undecenal | 1367 | nt | - | - | - | - | 0.1 ± 0.14 | - |
| α-copaene | 1376 | sh | - | - | - | 0.2 ± 0.04 | 0.3 ± 0.09 | 0.2 ± 0.01 |
| β-elemene | 1392 | sh | 0.2 ± 0.01 | 0.4 ± 0.07 | 0.3 ± 0.16 | 0.4 ± 0.15 | 0.4 ± 0.28 | 0.5 ± 0.28 |
| methyl eugenol | 1405 | pp | 3.7 ± 0.02 | 7.0 ± 1.28 | 1.8 ± 0.42 | 10.7 ± 5.15 | 15.4 ± 5.79 | 2.0 ± 0.45 |
| cis-α-bergamotene | 1416 | sh | - | - | - | - | 0.2 ± 0.03 | - |
| β-caryophyllene | 1419 | sh | - | 0.1 ± 0.02 | - | 0.1 ± 0.04 | 0.2 ± 0.10 | 0.2 ± 0.03 |
| trans-α-bergamotene | 1436 | sh | 5.8 ± 0.03 | 8.8 ± 0.69 | 8.5 ± 2.70 | 12 ± 3.92 | 16.5 ± 5.13 | 12.6 ± 2.32 |
| α-guaiene | 1439 | sh | 0.2 ± 0.01 | 0.2 ± 0.02 | - | 0.2 ± 0.12 | 0.3 ± 0.14 | 0.4 ± 0.09 |
| α-humulene | 1453 | sh | 0.2 ± 0.02 | 0.3 ± 0.06 | 0.1 ± 0.01 | 0.3 ± 0.07 | 0.4 ± 0.17 | 0.4 ± 0.05 |
| (E)-β-farnesene | 1458 | sh | 0.4 ± 0.05 | 0.6 ± 0.22 | 0.4 ± 0.13 | 1.3 ± 0.16 | 0.4 ± 0.16 | 0.6 ± 0.42 |
| germacrene D | 1481 | sh | 0.6 ± 0.00 | 0.7 ± 0.1 | 0.6 ± 0.33 | 0.9 ± 0.18 | 1.3 ± 0.47 | 0.9 ± 0.25 |
| trans-β-bergamotene | 1492 | sh | 0.2 ± 0.00 | 0.3 ± 0.04 | 0.3 ± 0.11 | 0.4 ± 0.12 | 0.5 ± 0.19 | 0.3 ± 0.12 |
| bicyclogermacrene | 1496 | sh | 0.1 ± 0.01 | 0.2 ± 0.01 | - | - | - | 0.2 ± 0.04 |
| α-bulnesene | 1505 | sh | 0.3 ± 0.01 | 0.3 ± 0.04 | 0.2 ± 0.11 | 0.3 ± 0.12 | 0.4 ± 0.23 | 0.5 ± 0.17 |
| trans-γ-cadinene | 1514 | sh | 0.2 ± 0.02 | 0.2 ± 0.04 | 0.3 ± 0.18 | 0.3 ± 0.12 | 0.3 ± 0.15 | 0.4 ± 0.02 |
| β-sesquiphellandrene | 1524 | sh | - | 0.2 ± 0.03 | 0.1 ± 0.08 | 0.2 ± 0.04 | 0.2 ± 0.10 | 0.2 ± 0.07 |
| T-cadinol | 1641 | os | - | 0.1 ± 0.01 | - | - | 0.2 ± 0.07 | |
| WT/CTR | 17A2 | 21D | 22B | 45B | 47C | |||
| Monoterpene hydrocarbons (mh) | 16.6 ± 0.15 B | 21.1 ± 2.01 AB | 24.6 ± 3.75 A | 20.3 ± 0.53 AB | 18.0 ± 0.77 B | 22.3 ± 3.46 AB | ||
| Oxygenated monoterpenes (om) | 64.5 ± 0.08 A | 39.4 ± 6.17 BC | 47.4 ± 6.75 B | 33.8 ± 3.56 C | 36.3 ± 4.41 BC | 47.1 ± 4.26 B | ||
| Sesquiterpenes hydrocarbons (sh) | 8.1 ± 0.05 B | 12.4 ± 1.30 AB | 10.8 ± 3.53 AB | 16.2 ± 4.96 AB | 21.4 ± 7.21 A | 17.2 ± 3.85 AB | ||
| Oxygenated sesquiterpenes (os) | - | 0.1 ± 0.01 AB | - | - | - | 0.2 ± 0.07 A | ||
| Phenylpropanoids (pp) | 10.7 ± 0.04 C | 27.0 ± 6.88 AB | 17.2 ± 6.96 ABC | 29.4 ± 8.1 A | 24.0 ± 3.38 ABC | 13.2 ± 3.93 BC | ||
| Other non-terpene derivatives (nt) | 0.1 ± 0.07 AB | - B | - B | - B | 0.1 ± 0.14 A | - B | ||
| Total identified (%) | 100.0 ± 0.01 | 100.0 ± 0.00 | 100.0 ± 0.00 | 99.6 ± 0.12 | 99.9 ± 0.06 | 100.0 ± 0.01 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laura, M.; Forti, C.; Barberini, S.; Ciorba, R.; Mascarello, C.; Giovannini, A.; Pistelli, L.; Pieracci, Y.; Lanteri, A.P.; Ronca, A.; et al. Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii. Plants 2023, 12, 2395. https://doi.org/10.3390/plants12132395
Laura M, Forti C, Barberini S, Ciorba R, Mascarello C, Giovannini A, Pistelli L, Pieracci Y, Lanteri AP, Ronca A, et al. Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii. Plants. 2023; 12(13):2395. https://doi.org/10.3390/plants12132395
Chicago/Turabian StyleLaura, Marina, Chiara Forti, Sara Barberini, Roberto Ciorba, Carlo Mascarello, Annalisa Giovannini, Luisa Pistelli, Ylenia Pieracci, Anna Paola Lanteri, Agostina Ronca, and et al. 2023. "Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii" Plants 12, no. 13: 2395. https://doi.org/10.3390/plants12132395
APA StyleLaura, M., Forti, C., Barberini, S., Ciorba, R., Mascarello, C., Giovannini, A., Pistelli, L., Pieracci, Y., Lanteri, A. P., Ronca, A., Minuto, A., Ruffoni, B., Cardi, T., & Savona, M. (2023). Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii. Plants, 12(13), 2395. https://doi.org/10.3390/plants12132395










