Analysis of Altered Flowering Related Genes in a Multi-Silique Rapeseed (Brassica napus L.) Line zws-ms Based on Combination of Genome, Transcriptome and Proteome Data
Abstract
1. Introduction
2. Results
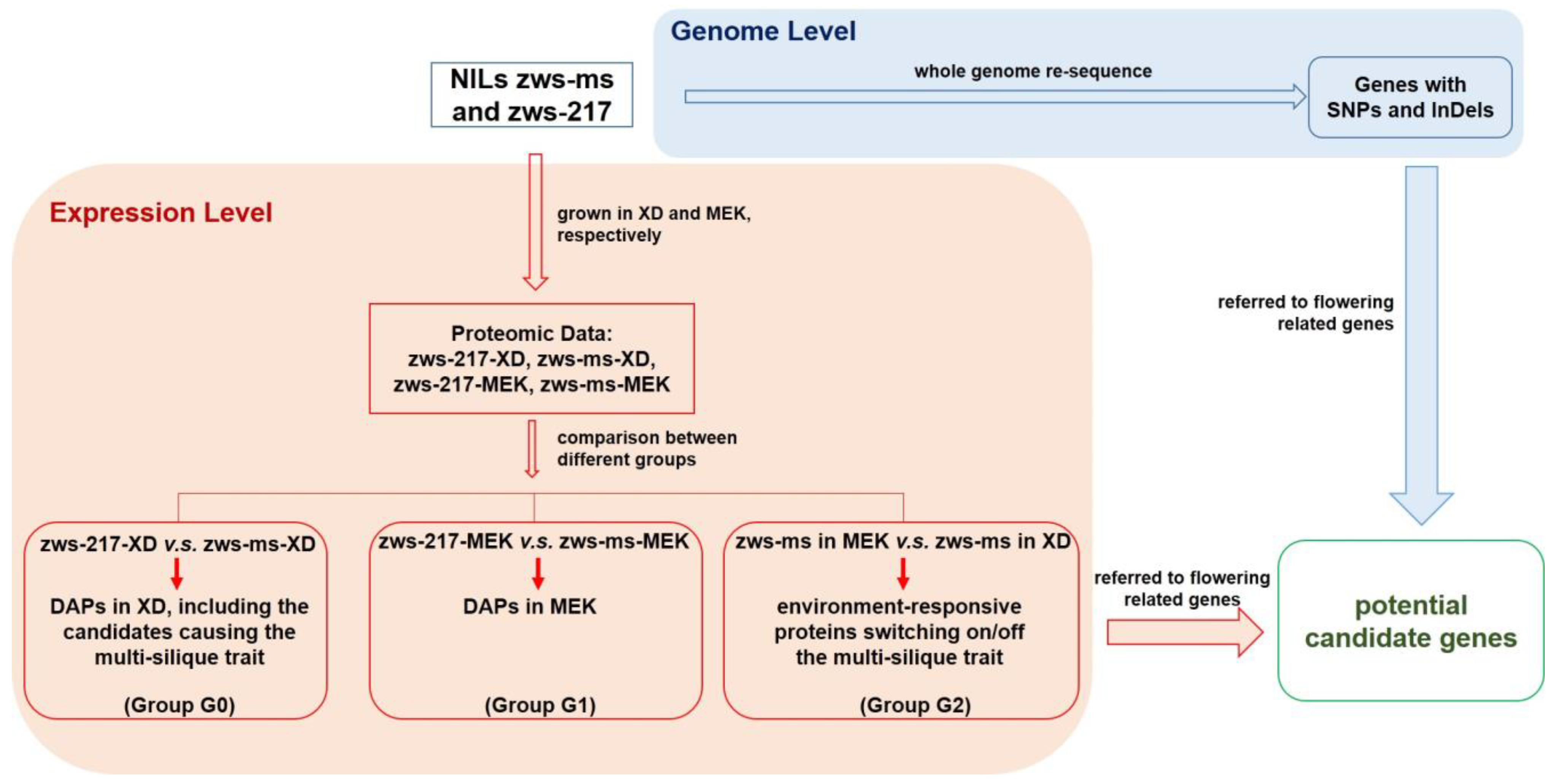
2.1. Identification of Differentially Accumulated Proteins between zws-ms and zws-217 in Two Environments
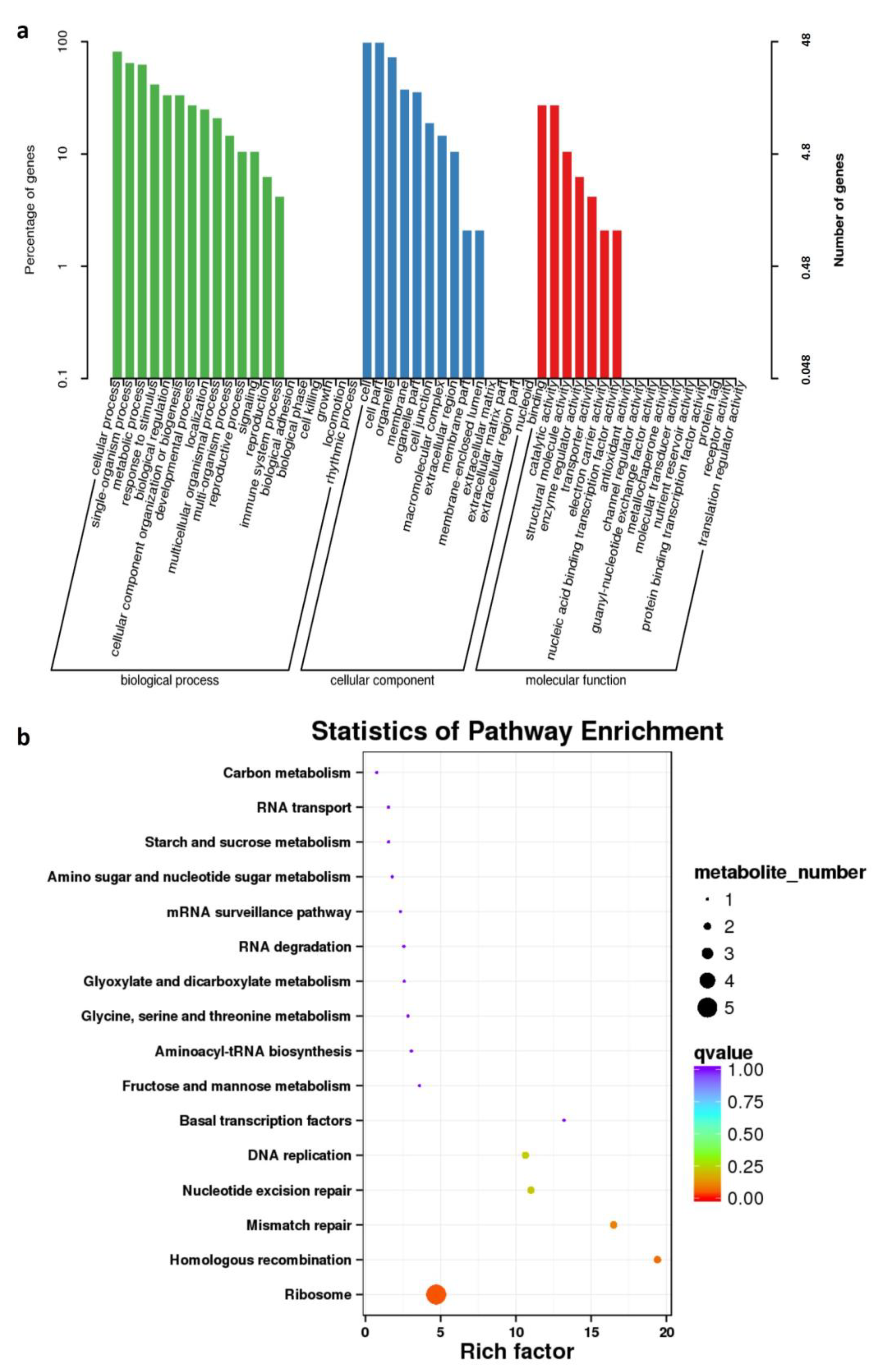
2.2. Comparison of DAPs among Different Groups and Their Annotations
2.3. Identification of the DAPs and DEGs Related to Flowering
2.4. Discovery of the Flowering-Realted Genes with Non-Synonymous SNP or Frame-Shift InDel in Line zws-ms
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. The Identification of the Flowering Related Genes in Rapeseed
4.3. Protein Extraction and Digestion
4.4. Liquid Chromatography Tandem Mass Spectrometry Analysis
4.5. Differentially Accumulated Proteins (DAPs) and Differentially Expressed Genes (DEGs) Identification
4.6. The Functional Annotations and Analysis of the Proteins
4.7. The Analysis of SNPs and InDels in the Flowering Related Genes on Genomic Level
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Seleiman, M.F.; Santanen, A.; Stoddard, F.L.; Mäkelä, P. Feedstock quality and growth of bioenergy crops fertilized with sewage sludge. Chemosphere 2012, 89, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, L.; Zhang, L.; Li, X.; Liu, Y.; Wu, Z.; Dong, F.; Wan, L.; Liu, K.; Hong, D. BnaC9. SMG7b functions as a positive regulator of the number of seeds per silique in Brassica napus by regulating the formation of functional female gametophytes. Plant Physiol. 2015, 169, 2744–2760. [Google Scholar] [PubMed]
- Chai, L.; Zhang, J.; Li, H.; Jiang, J.; Cui, C.; Zheng, B.; He, P.; Zhu, L.; Jiang, L. Dynamic Transcriptome Analysis of a Multi-Silique Trait in Rapeseed (Brassica napus L.). Int. J. Agric. Biol. 2020, 24, 1625–1632. [Google Scholar]
- Chai, L.; Zhang, J.; Li, H.; Zheng, B.; Jiang, J.; Cui, C.; Jiang, L. Investigation for a multi-silique trait in Brassica napus by alternative splicing analysis. PeerJ 2020, 8, e10135. [Google Scholar] [CrossRef]
- Chai, L.; Zhang, J.; Lu, K.; Li, H.; Wu, L.; Wan, H.; Zheng, B.; Cui, C.; Jiang, J.; Jiang, L. Identification of genomic regions associated with multi-silique trait in Brassica napus. BMC Genom. 2019, 20, 304. [Google Scholar] [CrossRef]
- Chai, L.; Zhang, J.; Li, H.; Cui, C.; Jiang, J.; Zheng, B.; Wu, L.; Jiang, L. Investigation of Thermomorphogenesis-Related Genes for a Multi-Silique Trait in Brassica napus by Comparative Transcriptome Analysis. Front. Genet. 2021, 12, 678804. [Google Scholar] [CrossRef]
- Zhu, X.; Ni, Y.; He, R.; Jiang, Y.; Li, Q.; Niu, J. Genetic mapping and expressivity of a wheat multi-pistil gene in mutant 12TP. J. Integr. Agric. 2019, 18, 532–538. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, G.; Song, Y.; Li, Z.; Ma, S.; Niu, N.; Wang, J. Comparative proteomic analysis of multi-ovary wheat under heterogeneous cytoplasm suppression. BMC Plant Biol. 2019, 19, 175. [Google Scholar] [CrossRef]
- Wei, S.H. Characterization and expression of WAG-2 transcripts in a wheat three-pistil mutant line. Russ. J. Plant Physl. 2017, 64, 680–687. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, Z.; Peng, Z.; Yu, Y.; Liao, M.; Wei, S. Development of a high-density linkage map and mapping of the three-pistil gene (Pis1) in wheat using GBS markers. BMC Genom. 2017, 18, 567. [Google Scholar] [CrossRef]
- Duan, Z.; Shen, C.; Li, Q.; Lü, G.; Ni, Y.; Yu, D.; Niu, J. Identification of a novel male sterile wheat mutant dms conferring dwarf status and multi-pistils. J. Integr. Agr. 2015, 14, 1706–1714. [Google Scholar] [CrossRef]
- Yang, Z.; Peng, Z.; Yang, H.; Yang, J.; Wei, S.; Cai, P. Suppression Subtractive Hybridization Identified Differentially Expressed Genes in Pistil Mutations in Wheat. Plant Mol. Biol. Rep. 2011, 29, 431–439. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, J.; Zhuang, H.; Zeng, X.; Tang, J.; Wang, H.; Chen, H.; Li, Y.; Ling, Y.; He, G.; et al. Gene mapping and candidate gene analysis of multi-floret spikelet 3 (mfs3) in rice (Oryza sativa L.). J. Integr. Agr. 2019, 18, 2673–2681. [Google Scholar] [CrossRef]
- Suzaki, T.; Sato, M.; Ashikari, M.; Miyoshi, M.; Nagato, Y.; Hirano, H. The gene Floral Organ Number1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; She, W.; Wang, L.; Luo, M.; Chen, Y.; Li, Y.; Wang, S.; Zhang, C. MADS-Box Genes are Involved in Cultivar- and Temperature-Dependent Formation of Multi-pistil and Polycarpy in Prunus avium L. J. Plant Growth Regul. 2019, 38, 1017–1027. [Google Scholar] [CrossRef]
- Wang, J.; Liu, J.; Jiu, S.; Li, Y.; Whiting, M.; She, W.; Wang, L.; Ma, C.; Xu, W.; Wang, S.; et al. The MADS-box genes PaMADS3/4/5 co-regulate multi-pistil formation induced by high temperature in Prunus avium L. Sci. Hortic. 2019, 256, 108593. [Google Scholar] [CrossRef]
- Liang, Y.; Gong, J.; Yan, Y.; Peng, T.; Xiao, J.; Wang, S.; Nan, W.; Qin, X.; Zhang, H. Fine Mapping and Candidate-Gene Analysis of an open glume multi-pistil 3 (mp3) in Rice (Oryza sativa L.). Agriculture 2022, 12, 1731. [Google Scholar] [CrossRef]
- Xu, W.; Tao, J.; Chen, M.; Dreni, L.; Luo, Z.; Hu, Y.; Liang, W.; Zhang, D. Interactions between Floral Organ Number4 and floral homeotic genes in regulating rice flower development. J. Exp. Bot. 2017, 68, 483–498. [Google Scholar] [CrossRef]
- Chu, H.; Qian, Q.; Liang, W.; Yin, C.; Tan, H.; Yao, X.; Yuan, Z.; Yang, J.; Huang, H.; Luo, D.; et al. The Floral Organ Number4 Gene Encoding a Putative Ortholog of Arabidopsis CLAVATA3 Regulates Apical Meristem Size in Rice. Plant Physiol. 2006, 142, 1039–1052. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Ma, L.; Sang, X.; Ling, Y.; Wang, Y.; Yu, P.; Zhuang, H.; Huang, J.; Wang, N.; et al. Lateral Floret 1 induced the three-florets spikelet in rice. Proc. Natl. Acad. Sci. USA 2017, 114, 9984–9989. [Google Scholar] [CrossRef]
- Peng, P.F.; Hu, Q.; Fu, L.; Chen, Y.F.; Liu, J.; Wang, H.; Sang, S.F.; Mei, D.S. Effect of BnFUL over-expression on flower and silique development in Arabidopsis thaliana. Chin. J. Oil Crop Sci. 2016, 38, 13–18. (In Chinese) [Google Scholar]
- Wang, Y.H.; Xie, L.; Yang, B.; Cao, Y.R.; Li, J.N. Flowering genes in oilseed rape: Identification, characterization, evolutionary and expression analysis. Acta Agron. Sin. 2019, 45, 1137–1145. (In Chinese) [Google Scholar]
- Wilkins, M.R.; Sanchez, J.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with Proteome Projects: Why all Proteins Expressed by a Genome Should be Identified and How to Do It. Biotechnol. Genet. Eng. Rev. 1996, 13, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Swinbanks, D. Government backs proteome proposal. Nature 1995, 378, 653. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Song, S.; Li, J.; Stewart, C.N.; Wei, W.; Zhao, Y.; Wang, W. A proteomic analysis of seeds from Bt-transgenic Brassica napus and hybrids with wild B. juncea. Sci. Rep. 2015, 5, 15480. [Google Scholar] [CrossRef]
- Cao, J.; Xu, Y.; Cai, X. TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia sclerotiorum. J. Proteom. 2016, 143, 265–277. [Google Scholar] [CrossRef]
- Jhanzab, H.; Razzaq, A.; Bibi, Y.; Yasmeen, F.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Komatsu, S. Proteomic Analysis of the Effect of Inorganic and Organic Chemicals on Silver Nanoparticles in Wheat. Int. J. Mol. Sci. 2019, 20, 825. [Google Scholar] [CrossRef]
- Wang, X.; Yang, R.; Zhou, Y.; Gu, Z. A comparative transcriptome and proteomics analysis reveals the positive effect of supplementary Ca2+ on soybean sprout yield and nutritional qualities. J. Proteom. 2016, 143, 161–172. [Google Scholar] [CrossRef]
- Gaeta, R.T.; Pires, J.C.; Iniguez-Luy, F.; Leon, E.; Osborn, T.C. Genomic Changes in Resynthesized Brassica napus and Their Effect on Gene Expression and Phenotype. Plant Cell 2007, 19, 3403–3417. [Google Scholar] [CrossRef]
- Xiong, Z.; Gaeta, R.T.; Pires, J.C. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 2011, 108, 7908–7913. [Google Scholar] [CrossRef]
- Leflon, M.; Grandont, L.; Eber, F.; Huteau, V.; Coriton, O.; Chelysheva, L.; Jenczewski, E.; Chèvre, A. Crossovers Get a Boost in Brassica Allotriploid and Allotetraploid Hybrids. Plant Cell 2010, 22, 2253–2264. [Google Scholar] [CrossRef]
- Ran, L.; Fang, T.; Rong, H.; Jiang, J.; Fang, Y.; Wang, Y. Analysis of cytosine methylation in early generations of resynthesized Brassica napus. J. Integr. Agric. 2016, 15, 1228–1238. [Google Scholar] [CrossRef]
- Steffen, A.; Elgner, M.; Staiger, D. Regulation of Flowering Time by the RNA-Binding Proteins AtGRP7 and AtGRP8. Plant Cell Physiol. 2019, 60, 2040–2050. [Google Scholar] [CrossRef]
- Wang, J.; Sun, W.; Wang, L.; Liu, X.; Xu, Y.; Sabir, I.A.; Jiu, S.; Wang, S.; Zhang, C. Fruitfull is involved in double fruit formation at high temperature in sweet cherry. Environ. Exp. Bot. 2022, 201, 104986. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative Regulation of the Shatterproof Genes by Fruitfull During Arabidopsis Fruit Development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef]
- Peng, P.F.; Li, Y.C.; Mei, D.S.; Liu, D.M.; Fu, L.; Wang, H.; Sang, S.F.; Chen, Y.F.; Hu, Q. Optimization and experiment of assessment method for pod shatter resistance in Brassica napus L. Trans. Chin. Soc. Agric. Eng. 2013, 29, 19–25. (In Chinese) [Google Scholar]
- Zhu, X.; Zhang, L.; Kuang, C.; Guo, Y.; Huang, C.; Deng, L.; Sun, X.; Zhan, G.; Hu, Z.; Wang, H.; et al. Important photosynthetic contribution of silique wall to seed yield-related traits in Arabidopsis thaliana. Photosynth. Res. 2018, 137, 493–501. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef]
- Proveniers, M.; Rutjens, B.; Brand, M.; Smeekens, S. The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. Plant J. 2007, 52, 899–913. [Google Scholar] [CrossRef]
- Yu, Z.Y.; Luo, Q.; Peng, Z.S.; Wei, S.H.; Yang, Z.J.; Yamamoto, N. Genetic mapping of the three-pistil gene Pis1 in an F2 population derived from a synthetic hexaploid wheat using multiple molecular marker systems. Cereal Res. Commun. 2021, 49, 31–36. [Google Scholar] [CrossRef]
- Guo, J.; Li, Z.; Zhang, G.; Tang, H.; Song, Q.; Song, Y.; Ma, S.; Niu, N.; Wang, J. Special heterogeneous cytoplasm suppresses the expression of the gene producing multi-ovary in common wheat. Euphytica 2017, 213, 247. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248. [Google Scholar] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Zeng, Y.; Liang, Y.; Qian, Q.; Yang, C. Proteomic dissection of the rice-Fusarium fujikuroi interaction and the correlation between the proteome and transcriptome under disease stress. BMC Genom. 2019, 20, 91. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, X.; Li, P.; Zhou, F.; Kong, L.; Qiu, J.; Yuan, Z.; Tan, J. TMT Based Proteomic Analysis of Human Follicular Fluid From Overweight/Obese and Normal-Weight Patients With Polycystic Ovary Syndrome. Front. Endocrinol. 2019, 10, 821. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]


| Comparison | Total DAPs | Up-Regulated DAPs in zws-ms | Down-Regulated DAPs in zws-ms |
|---|---|---|---|
| zws-217 v.s. zws-ms in Xindu (G0) | 66 | 41 | 25 |
| zws-217 v.s. zws-ms in Ma’erkang (G1) | 153 | 90 | 63 |
| zws-ms in Ma’erkang v.s. zws-ms in Xindu (G2) | 214 | 77 | 137 |
| Gene ID | Position | Ref | zws-ms | zws-217 | Swissprot Annotation | Ortholog in Arabidopsis | |
|---|---|---|---|---|---|---|---|
| Gene ID | Description | ||||||
| BnaA09g05900D | 2,880,166 | C | Y (C/T) | C | MADS-box protein AGL42 | AT5G62165 | AGAMOUS-like 42 (AGL42) |
| BnaC02g42040D | 44,852,629 | G | G | R (A/G) | Protein LEO1 homolog | AT5G61150 | VERNALIZATION INDEPENDENCE 4 (VIP4) |
| BnaC08g36330D | 33,648,610 | A | R (A/G) | A | Transcription activator GLK1 | AT2G20570 | GBF’s pro-rich region-interacting factor 1 (GPRI1) |
| 33,648,961 | A | G | R (A/G) | ||||
| BnaA09g57260D | 4,114,551 | A | A | R (A/G) | Transcription factor bHLH74 | AT1G10120 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Gene ID | Pos | Ref | zws-ms | zws-217 | Swissprot Annotation | Ortholog in Arabidopsis | |
|---|---|---|---|---|---|---|---|
| Gene ID | Gene ID | ||||||
| BnaA02g28220D | 20,796,604 | G | G,GAGAGGGAGAGCA | G | Transcription factor ICE1 | AT3G26744 | INDUCER OF CBF EXPRESSION 1 (ICE1) |
| 20,796,652 | A | A,AACC | A | ||||
| 20,796,686 | C | C,CTGTAACATG | C | ||||
| 20,797,054 | CTT | CTT | CTT,C | ||||
| BnaA09g57260D | 4,114,345 | G | GTAA | G,GTAA | Transcription factor bHLH74 | AT1G10120 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| 4,114,386 | GT | G | GT,G | ||||
| 4,115,144 | AT | AT | AT,A | ||||
| 4,115,529 | C | C | C,CCCA | ||||
| BnaAnng33220D | 37,892,579 | C | CGTCTCCTCAAGGG | C,CGTCTCCTCAAGGG | WD-40 repeat-containing protein MSI4 | AT2G19520 | FVE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, L.; Li, H.; Zhao, X.; Cui, C.; Zheng, B.; Zhang, K.; Jiang, J.; Zhang, J.; Jiang, L. Analysis of Altered Flowering Related Genes in a Multi-Silique Rapeseed (Brassica napus L.) Line zws-ms Based on Combination of Genome, Transcriptome and Proteome Data. Plants 2023, 12, 2429. https://doi.org/10.3390/plants12132429
Chai L, Li H, Zhao X, Cui C, Zheng B, Zhang K, Jiang J, Zhang J, Jiang L. Analysis of Altered Flowering Related Genes in a Multi-Silique Rapeseed (Brassica napus L.) Line zws-ms Based on Combination of Genome, Transcriptome and Proteome Data. Plants. 2023; 12(13):2429. https://doi.org/10.3390/plants12132429
Chicago/Turabian StyleChai, Liang, Haojie Li, Xiaoguang Zhao, Cheng Cui, Benchuan Zheng, Ka Zhang, Jun Jiang, Jinfang Zhang, and Liangcai Jiang. 2023. "Analysis of Altered Flowering Related Genes in a Multi-Silique Rapeseed (Brassica napus L.) Line zws-ms Based on Combination of Genome, Transcriptome and Proteome Data" Plants 12, no. 13: 2429. https://doi.org/10.3390/plants12132429
APA StyleChai, L., Li, H., Zhao, X., Cui, C., Zheng, B., Zhang, K., Jiang, J., Zhang, J., & Jiang, L. (2023). Analysis of Altered Flowering Related Genes in a Multi-Silique Rapeseed (Brassica napus L.) Line zws-ms Based on Combination of Genome, Transcriptome and Proteome Data. Plants, 12(13), 2429. https://doi.org/10.3390/plants12132429





