ABA Biosynthesis- and Signaling-Related Gene Expression Differences between Sweet Cherry Fruits Suggest Attenuation of ABA Pathway in Bicolored Cultivars
Abstract
:1. Introduction
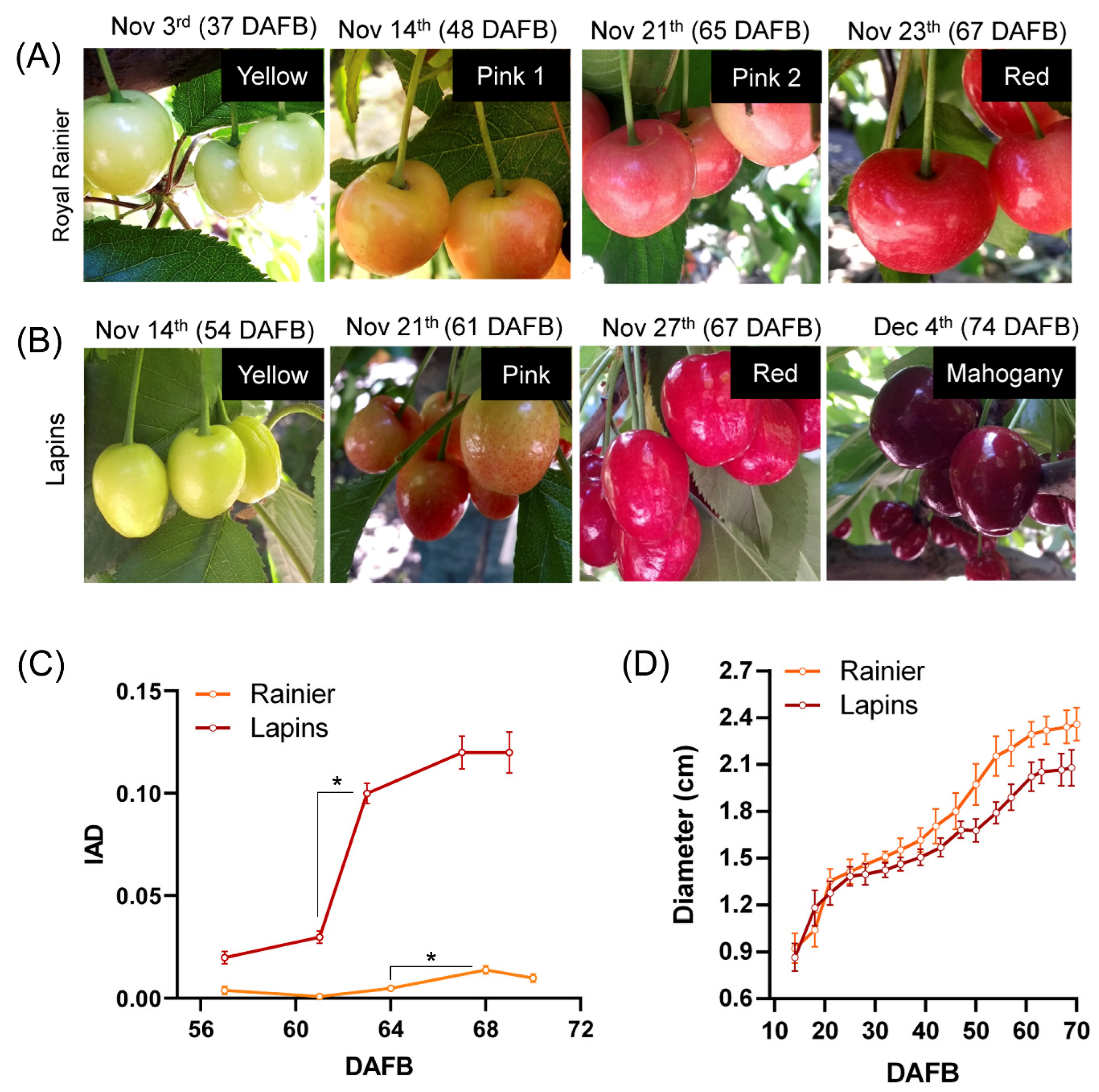
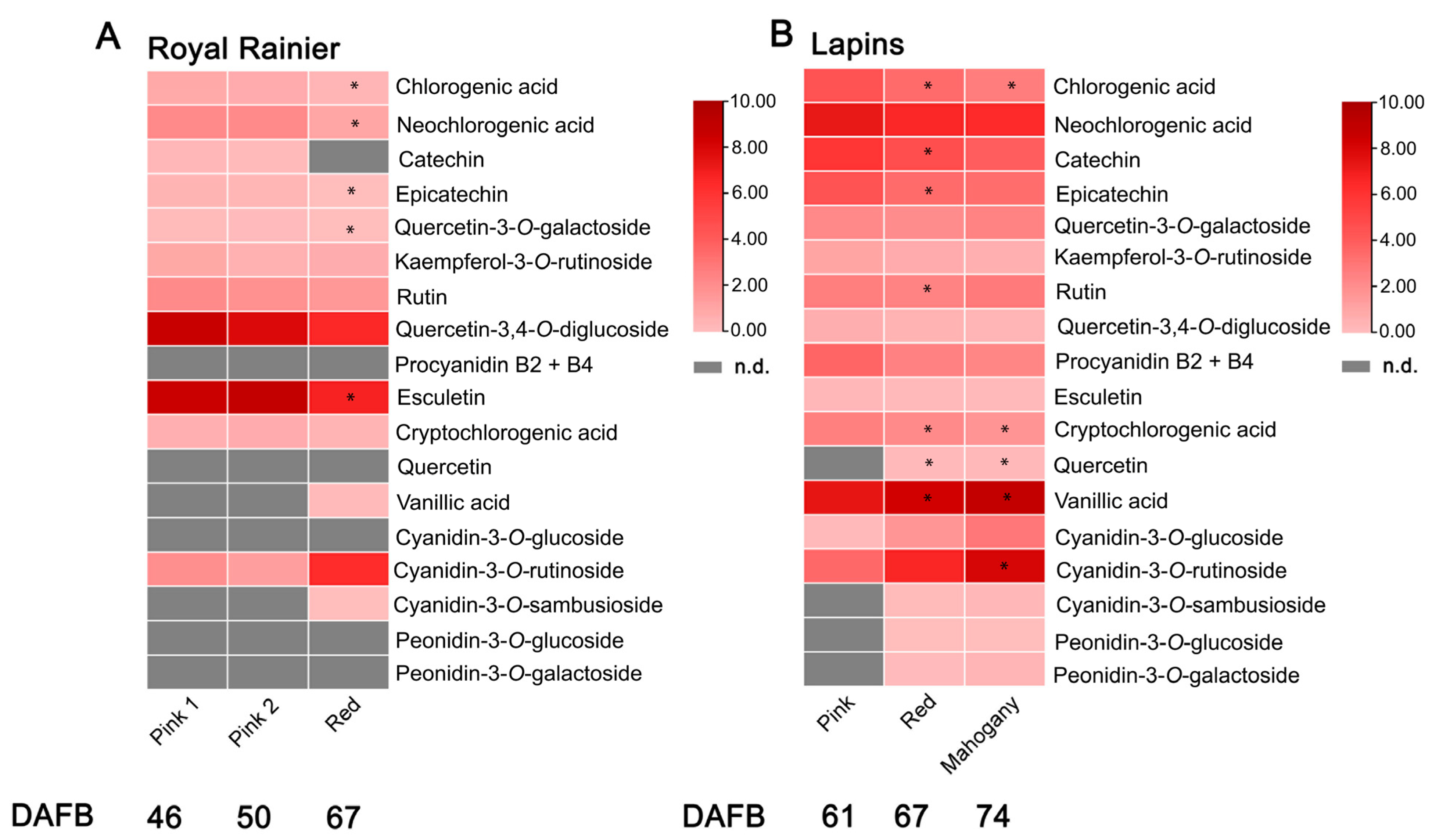
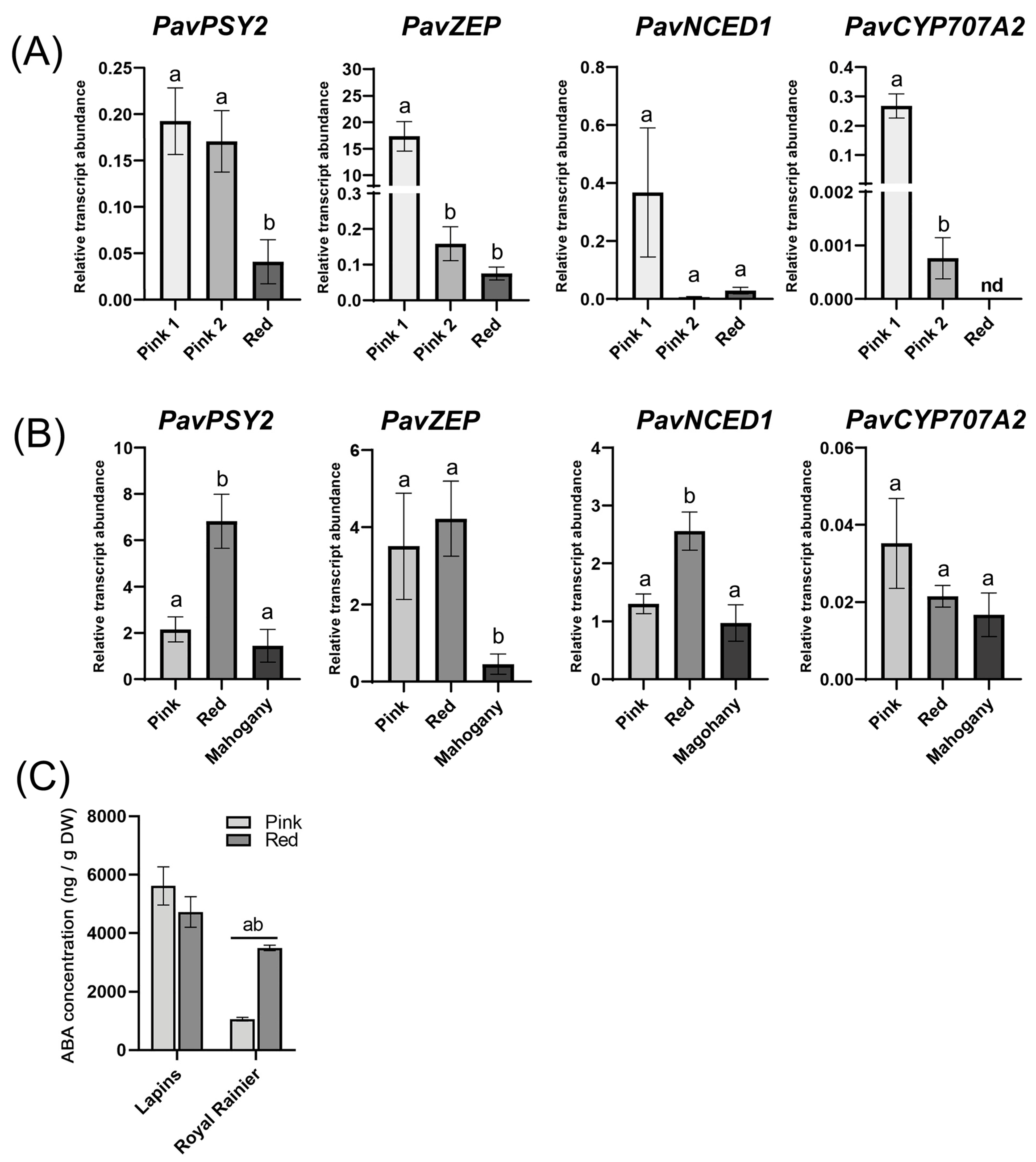
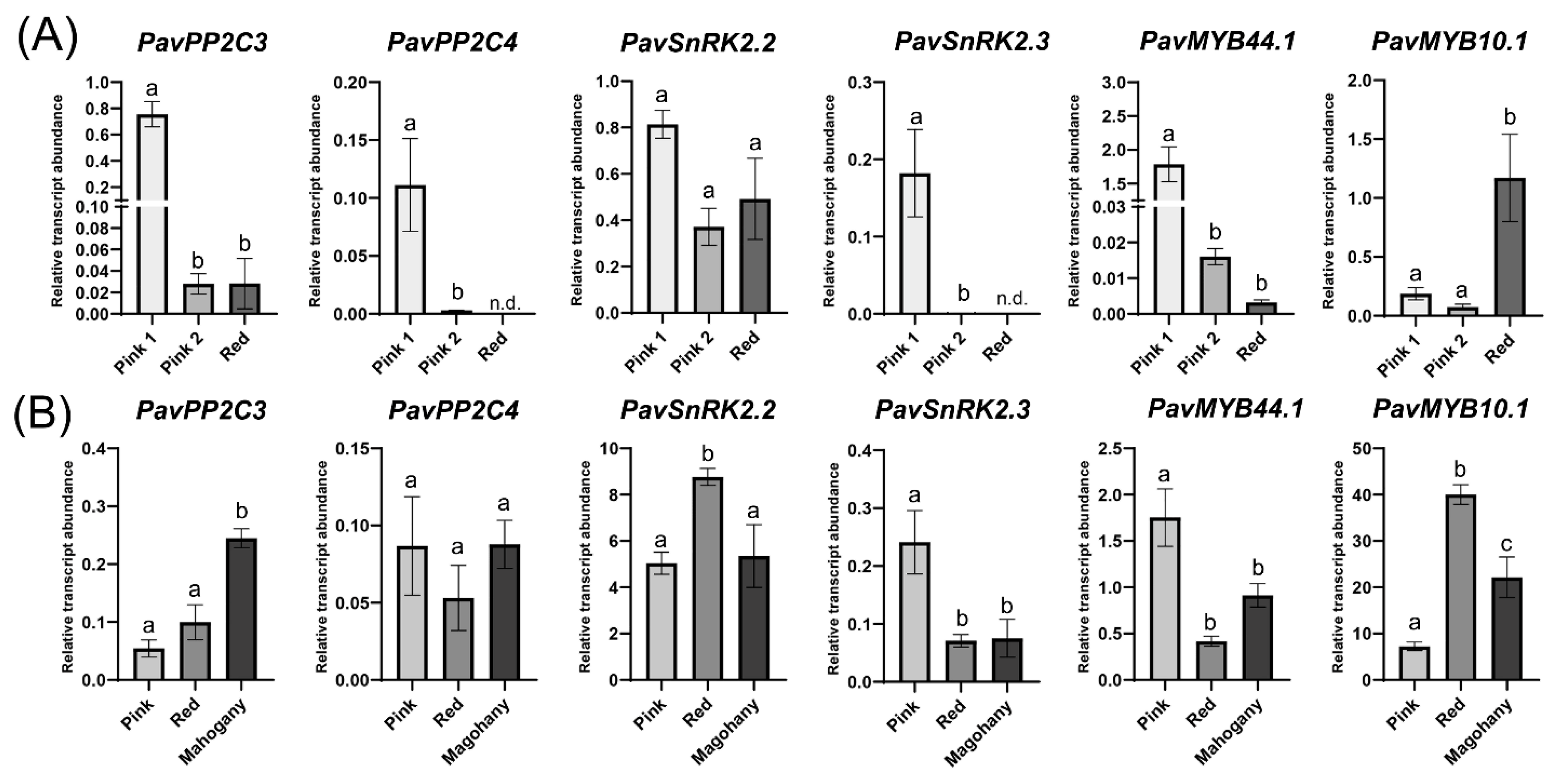
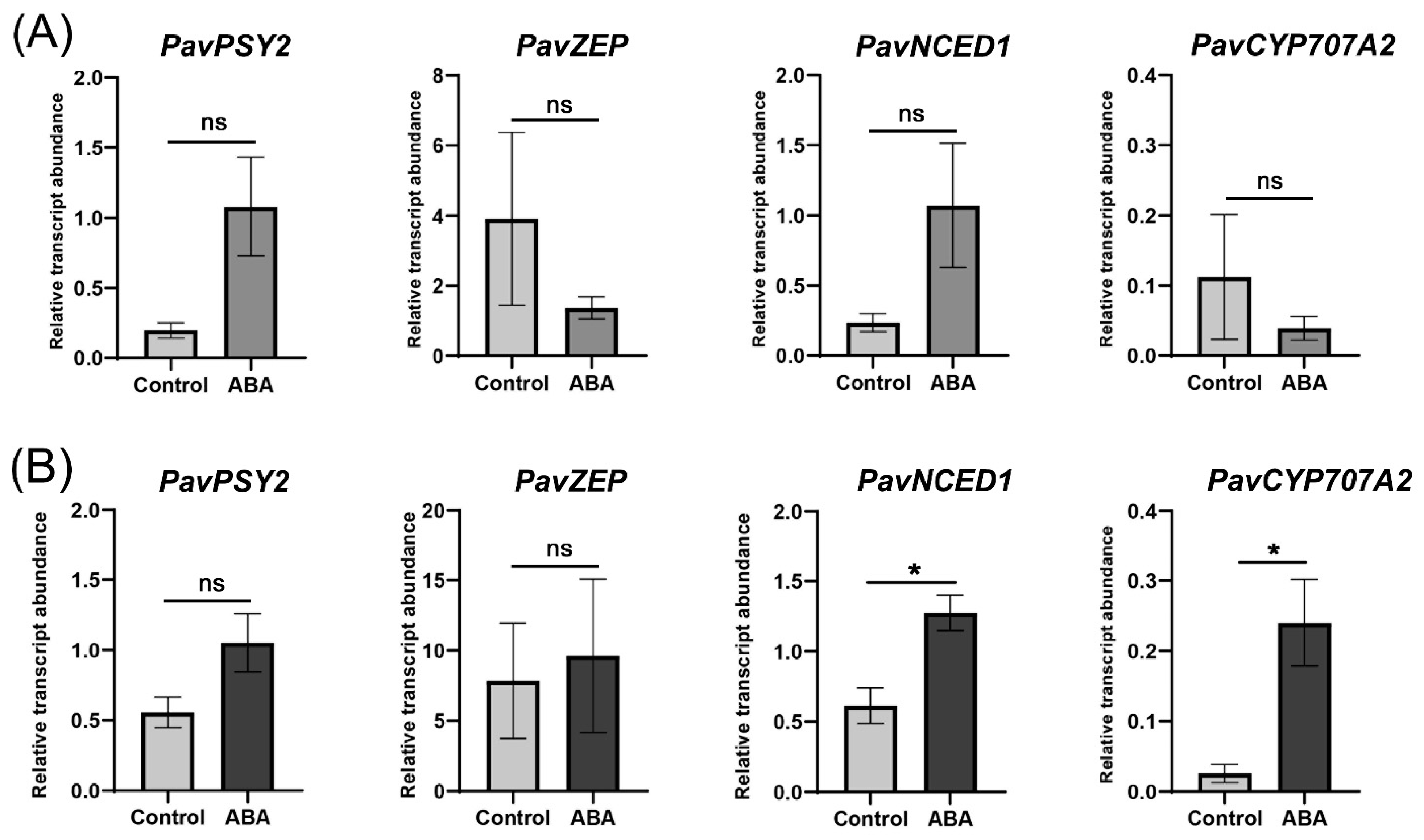
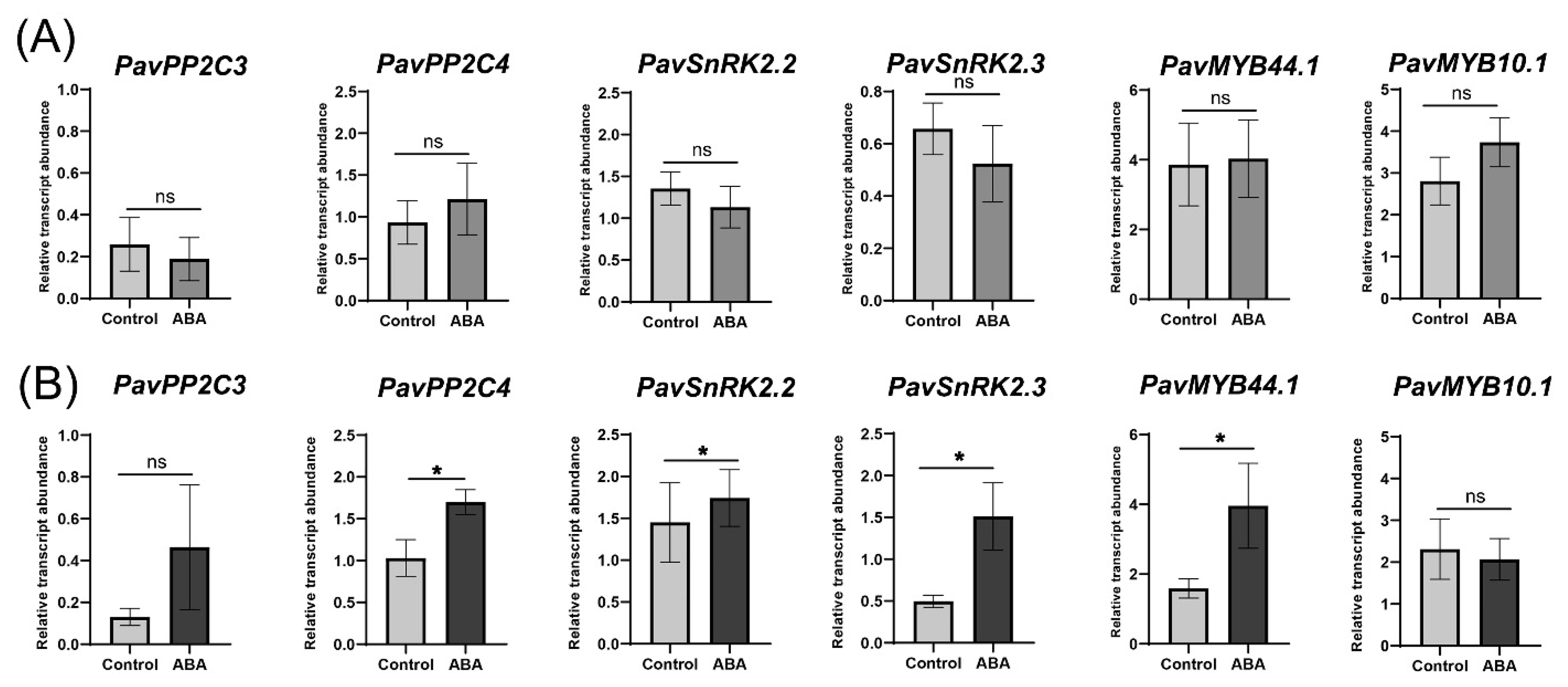
2. Results
3. Methods
3.1. Plant Material
3.2. ABA Treatment
3.3. Physiological Evaluations
3.4. Targeted Metabolomics of Phenolic Compounds
3.5. Hormone Quantification
3.6. RNA Extraction, cDNA Synthesis and RT-qPCR Analyses
3.7. In Silico Analyses of UTRs
4. Discussion
4.1. Differences between Dark-Red and Bicolored Fruits at the Metabolic and Gene Expression Level
4.2. Dark-Red and Bicolored Fruits Respond Differently to ABA
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, P.; Kumari, A.; Sheoran, B.; Sharma, S.; Kaur, S.; Bhunia, R.K.; Rajarammohan, S.; Bishnoi, M.; Kondepudi, K.K.; Garg, M. Anthocyanin Biofortified Colored Wheat Modifies Gut Microbiota in Mice. J. Cereal Sci. 2022, 104, 103433. [Google Scholar] [CrossRef]
- Jezek, M.; Zörb, C.; Merkt, N.; Geilfus, C.-M. Anthocyanin Management in Fruits by Fertilization. J. Agric. Food Chem. 2018, 66, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, N.; Luo, Z.; Yang, M.; Liu, Z.; Liu, M. The Underlying Molecular Mechanisms of External Factors Influencing Fruit Coloration in Fruit Trees. Sci. Hortic. 2023, 309, 111615. [Google Scholar] [CrossRef]
- Kayesh, E.; Shangguan, L.; Korir, N.K.; Sun, X.; Bilkish, N.; Zhang, Y.; Han, J.; Song, C.; Cheng, Z.-M.; Fang, J. Fruit Skin Color and the Role of Anthocyanin. Acta Physiol. Plant. 2013, 35, 2879–2890. [Google Scholar] [CrossRef]
- Hama, J.R.; Hooshmand, K.; Laursen, B.B.; Vestergård, M.; Fomsgaard, I.S. Clover Root Uptake of Cereal Benzoxazinoids (BXs) Caused Accumulation of BXs and BX Transformation Products Concurrently with Substantial Increments in Clover Flavonoids and Abscisic Acid. J. Agric. Food Chem. 2022, 70, 14633–14640. [Google Scholar] [CrossRef]
- Ferrer, J.-L.; Austin, M.B.; Stewart, C., Jr.; Noel, J.P. Structure and Function of Enzymes Involved in the Biosynthesis of Phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Rastogi, S.; Dwivedi, U.N. Phenylpropanoid Metabolism in Ripening Fruits. Compr. Rev. Food Sci. Food Saf. 2010, 9, 398–416. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, X.; Zhao, K.; Ben, Y.; Guo, X.; Zhang, X.; Li, T. Expression Analysis of Anthocyanin Biosynthetic Genes in Different Colored Sweet Cherries (Prunus avium L.) during Fruit Development. J. Plant Growth Regul. 2013, 32, 901–907. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB Transcription Factor PavMYB10.1 Involves in Anthocyanin Biosynthesis and Determines Fruit Skin Colour in Sweet Cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [Green Version]
- Medda, S.; Sanchez-Ballesta, M.T.; Romero, I.; Dessena, L.; Mulas, M. Expression of Structural Flavonoid Biosynthesis Genes in Dark-Blue and White Myrtle Berries (Myrtus Communis L.). Plants 2021, 10, 316. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB Transcription Factors That Colour Our Fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, N.; Ponce, C.; Arellano, M.; Time, A.; Multari, S.; Martens, S.; Carrera, E.; Sagredo, B.; Donoso, J.M.; Meisel, L.A. ABA Influences Color Initiation Timing in P. avium L. Fruits by Sequentially Modulating the Transcript Levels of ABA and Anthocyanin-Related Genes. Tree Genet. Genomes 2021, 17, 20. [Google Scholar] [CrossRef]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple Skin Patterning Is Associated with Differential Expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White Grapes Arose through the Mutation of Two Similar and Adjacent Regulatory Genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.; Loveys, B.; Ford, C.; Davies, C. The Relationship between the Expression of Abscisic Acid Biosynthesis Genes, Accumulation of Abscisic Acid and the Promotion of Vitis vinifera L. Berry Ripening by Abscisic Acid. Aust. J. Grape Wine Res. 2009, 15, 195–204. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.; Li, F.; Li, T. A Role for PacMYBA in ABA-Regulated Anthocyanin Biosynthesis in Red-Colored Sweet Cherry Cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Gupta, K.; Wani, S.H.; Razzaq, A.; Skalicky, M.; Samantara, K.; Gupta, S.; Pandita, D.; Goel, S.; Grewal, S.; Hejnak, V.; et al. Abscisic Acid: Role in Fruit Development and Ripening. Front. Plant Sci. 2022, 13, 817500. [Google Scholar] [CrossRef]
- Kadomura-Ishikawa, Y.; Miyawaki, K.; Takahashi, A.; Masuda, T.; Noji, S. Light and Abscisic Acid Independently Regulated FaMYB10 in Fragaria × Ananassa Fruit. Planta 2015, 241, 953–965. [Google Scholar] [CrossRef]
- Teribia, N.; Tijero, V.; Munné-Bosch, S. Linking Hormonal Profiles with Variations in Sugar and Anthocyanin Contents during the Natural Development and Ripening of Sweet Cherries. New Biotechnol. 2016, 33, 824–833. [Google Scholar] [CrossRef]
- Ponce, C.; Kuhn, N.; Arellano, M.; Time, A.; Multari, S.; Martens, S.; Carrera, E.; Sagredo, B.; Donoso, J.M.; Meisel, L.A. Differential Phenolic Compounds and Hormone Accumulation Patterns between Early- and Mid-Maturing Sweet Cherry (Prunus avium L.) Cultivars during Fruit Development and Ripening. J. Agric. Food Chem. 2021, 69, 8850–8860. [Google Scholar] [CrossRef]
- Luo, H.; Dai, S.; Ren, J.; Zhang, C.; Ding, Y.; Li, Z.; Sun, Y.; Ji, K.; Wang, Y.; Li, Q.; et al. The Role of ABA in the Maturation and Postharvest Life of a Nonclimacteric Sweet Cherry Fruit. J. Plant Growth Regul. 2014, 33, 373–383. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB Transcription Factor Associated with Regulation of the Anthocyanin Biosynthetic Pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Starkevič, P.; Paukštytė, J.; Kazanavičiūtė, V.; Denkovskienė, E.; Stanys, V.; Bendokas, V.; Šikšnianas, T.; Ražanskienė, A.; Ražanskas, R. Expression and Anthocyanin Biosynthesis-Modulating Potential of Sweet Cherry (Prunus avium L.) MYB10 and BHLH Genes. PLoS ONE 2015, 10, e0126991. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Zhai, Z.; Huang, T.; Zhao, D.; Peng, X.; Feng, C.; Xiao, Y.; Li, T. Transcriptomic Analysis of Light-Dependent Anthocyanin Accumulation in Bicolored Cherry Fruits. Plant Physiol. Biochem. 2018, 130, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Loyola, R.; Vega, A.; Peña-Neira, A.; Bordeu, E.; Arce-Johnson, P.; Alcalde, J.A. Post-Veraison Sunlight Exposure Induces MYB-Mediated Transcriptional Regulation of Anthocyanin and Flavonol Synthesis in Berry Skins of Vitis vinifera. J. Exp. Bot. 2009, 60, 853–867. [Google Scholar] [CrossRef] [Green Version]
- Mattus-Araya, E.; Guajardo, J.; Herrera, R.; Moya-León, M.A. ABA Speeds up the Progress of Color in Developing F. Chiloensis Fruit through the Activation of PAL, CHS and ANS, Key Genes of the Phenylpropanoid/Flavonoid and Anthocyanin Pathways. Int. J. Mol. Sci. 2022, 23, 3854. [Google Scholar] [CrossRef] [PubMed]
- Vrhovsek, U.; Masuero, D.; Gasperotti, M.; Franceschi, P.; Caputi, L.; Viola, R.; Mattivi, F. A Versatile Targeted Metabolomics Method for the Rapid Quantification of Multiple Classes of Phenolics in Fruits and Beverages. J. Agric. Food Chem. 2012, 60, 8831–8840. [Google Scholar] [CrossRef] [PubMed]
- Arapitsas, P.; Perenzoni, D.; Nicolini, G.; Mattivi, F. Study of Sangiovese Wines Pigment Profile by UHPLC-MS/MS. J. Agric. Food Chem. 2012, 60, 10461–10471. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Meisel, L.; Fonseca, B.; González, S.; Baeza-Yates, R.; Cambiazo, V.; Campos, R.; Gonźalez, M.; Orellana, A.; Retamales, J.; Silva, H. A Rapid and Efficient Method for Purifying High Quality Total RNA from Peaches (Prunus Persica) for Functional Genomics Analyses. Biol. Res. 2005, 38, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Alkio, M.; Jonas, U.; Declercq, M.; Van Nocker, S.; Knoche, M. Transcriptional Dynamics of the Developing Sweet Cherry (Prunus avium L.) Fruit: Sequencing, Annotation and Expression Profiling of Exocarp-Associated Genes. Hortic. Res. 2014, 1, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Chen, P.; Dai, S.; Sun, Y.; Yuan, B.; Kai, W.; Pei, Y.; He, S.; Liang, B.; Zhang, Y.; et al. PacCYP707A2 Negatively Regulates Cherry Fruit Ripening While PacCYP707A1 Mediates Drought Tolerance. J. Exp. Bot. 2015, 66, 3765–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, P.; Sun, L.; Li, Q.; Dai, S.; Sun, Y.; Kai, W.; Zhang, Y.; Liang, B.; Leng, P. Transcriptional Regulation of PaPYLs, PaPP2Cs and PaSnRK2s during Sweet Cherry Fruit Development and in Response to Abscisic Acid and Auxin at Onset of Fruit Ripening. Plant Growth Regul. 2015, 75, 455–464. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The Genome Sequence of Sweet Cherry (Prunus avium) for Use in Genomics-Assisted Breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification Efficiency: Linking Baseline and Bias in the Analysis of Quantitative PCR Data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.-R. Eleven Golden Rules of Quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Jia, C.; Waterhouse, G.I.N.; Sun-Waterhouse, D.; Sun, Y.G.; Wu, P. Variety–Compound–Quality Relationship of 12 Sweet Cherry Varieties ByHPLC-chemometric Analysis. Int. J. Food Sci. Technol. 2019, 54, 2897–2914. [Google Scholar] [CrossRef]
- Kobayashi, S.; Goto-Yamamoto, N.; Hirochika, H. Retrotransposon-Induced Mutations in Grape Skin Color. Science 2004, 304, 982. [Google Scholar] [CrossRef]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic Acid Plays an Important Role in the Regulation of Strawberry Fruit Ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Time, A.; Ponce, C.; Kuhn, N.; Arellano, M.; Sagredo, B.; Donoso, J.M.; Meisel, L.A. Canopy Spraying of Abscisic Acid to Improve Fruit Quality of Different Sweet Cherry Cultivars. Agronomy 2021, 11, 1947. [Google Scholar] [CrossRef]



| ‘Royal Rainier’ (mg/100 g FW) | ‘Lapins’ (mg/100 g FW) | |||||
|---|---|---|---|---|---|---|
| Pink 1 | Pink 2 | Red | Pink | Red | Mahogany | |
| Cyanidin-3-O-glucoside | n.d. | n.d. | n.d. | 0.08 ± 0.02 a | 2.44 ± 0.48 b | 8.51 ± 0.39 c |
| Cyanidin-3-O-rutinoside | 2.89 ± 0.72 a | 1.47 ± 0.45 a | 86.2 ± 45.22 a | 13.98 ± 2.5 a | 178.56 ± 32.04 b | 521.56 ± 20.33 c |
| Cyanidin-3-O-Sambubioside | n.d. | n.d. | 0.05 ± 0.04 | n.d. | 0.04 ± 0.01 a | 0.17 ± 0.01 b |
| Peonidin-3-O-glucoside | n.d. | n.d. | n.d. | n.d. | <0.01 a | <0.01 b |
| Peonidin-3-O-galactoside | n.d. | n.d. | n.d. | n.d. | 0.02 ± 0.01 a | 0.23 ± 0.12 a |
| Total Anthocyanins | 2.89 ± 0.72 a | 1.47 ± 0.45 a | 86.25 ± 45.26 a | 14.06 ± 2.6 a | 181.24 ± 32.54 b | 530.47076 ± 20.85 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acevedo, O.; Ponce, C.; Arellano, M.; Multari, S.; Carrera, E.; Donoso, J.M.; Martens, S.; Kuhn, N.; Meisel, L.A. ABA Biosynthesis- and Signaling-Related Gene Expression Differences between Sweet Cherry Fruits Suggest Attenuation of ABA Pathway in Bicolored Cultivars. Plants 2023, 12, 2493. https://doi.org/10.3390/plants12132493
Acevedo O, Ponce C, Arellano M, Multari S, Carrera E, Donoso JM, Martens S, Kuhn N, Meisel LA. ABA Biosynthesis- and Signaling-Related Gene Expression Differences between Sweet Cherry Fruits Suggest Attenuation of ABA Pathway in Bicolored Cultivars. Plants. 2023; 12(13):2493. https://doi.org/10.3390/plants12132493
Chicago/Turabian StyleAcevedo, Orlando, Claudio Ponce, Macarena Arellano, Salvatore Multari, Esther Carrera, José Manuel Donoso, Stefan Martens, Nathalie Kuhn, and Lee A. Meisel. 2023. "ABA Biosynthesis- and Signaling-Related Gene Expression Differences between Sweet Cherry Fruits Suggest Attenuation of ABA Pathway in Bicolored Cultivars" Plants 12, no. 13: 2493. https://doi.org/10.3390/plants12132493
APA StyleAcevedo, O., Ponce, C., Arellano, M., Multari, S., Carrera, E., Donoso, J. M., Martens, S., Kuhn, N., & Meisel, L. A. (2023). ABA Biosynthesis- and Signaling-Related Gene Expression Differences between Sweet Cherry Fruits Suggest Attenuation of ABA Pathway in Bicolored Cultivars. Plants, 12(13), 2493. https://doi.org/10.3390/plants12132493








