Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Aluminum, Copper and Cadmium Accumulation in Tea ‘Qianmei 419’ and ‘Qianfu 4’
Abstract
:1. Introduction
2. Results
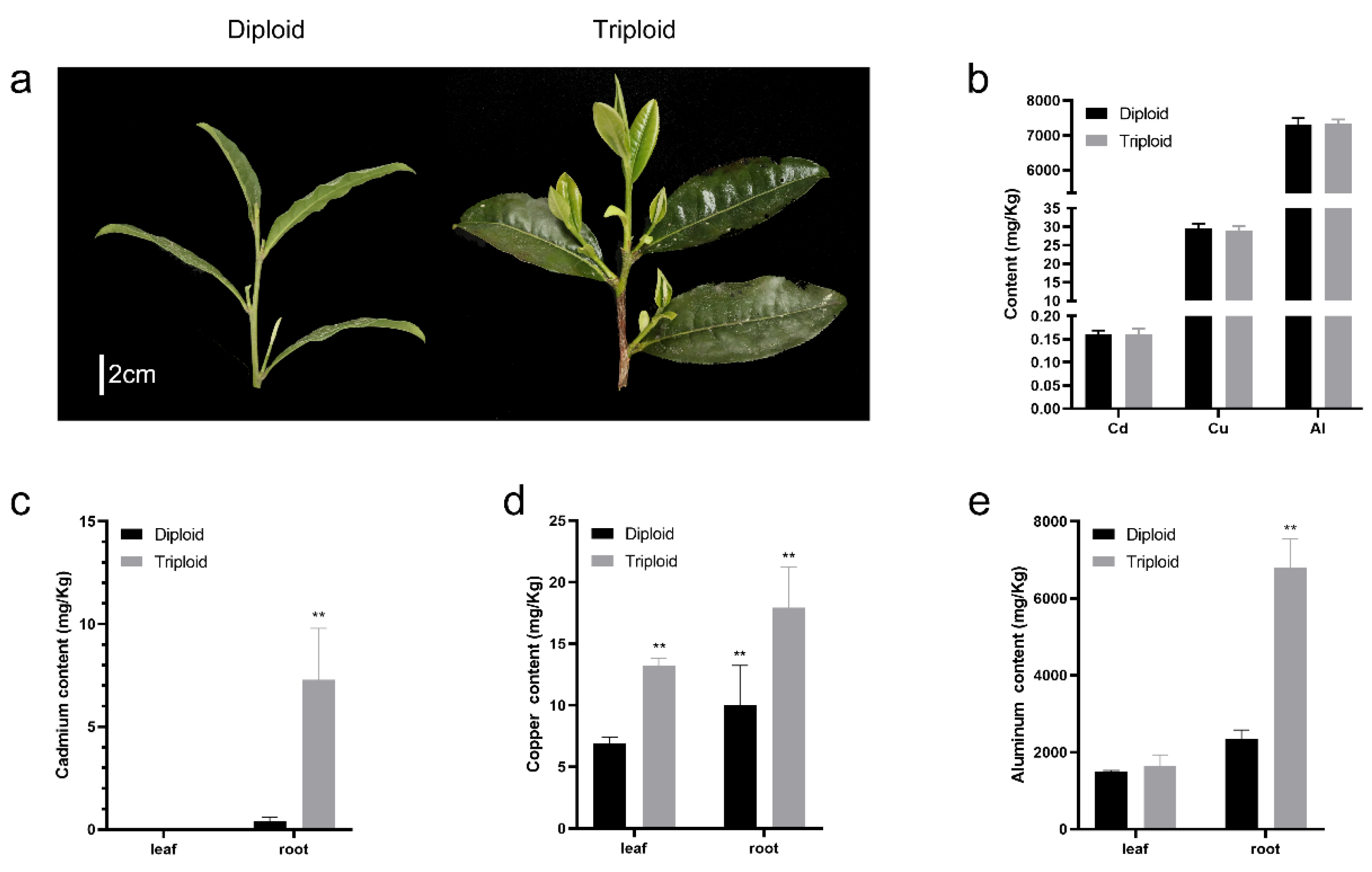
2.1. Measurements of Al, Cd, and Cu Contents in ‘Qianmei 419’ and ‘QianFu No.4’
2.2. RNA-Seq Analysis of ‘Qianmei 419’ and ‘QianFu No.4’ Tea Varieties
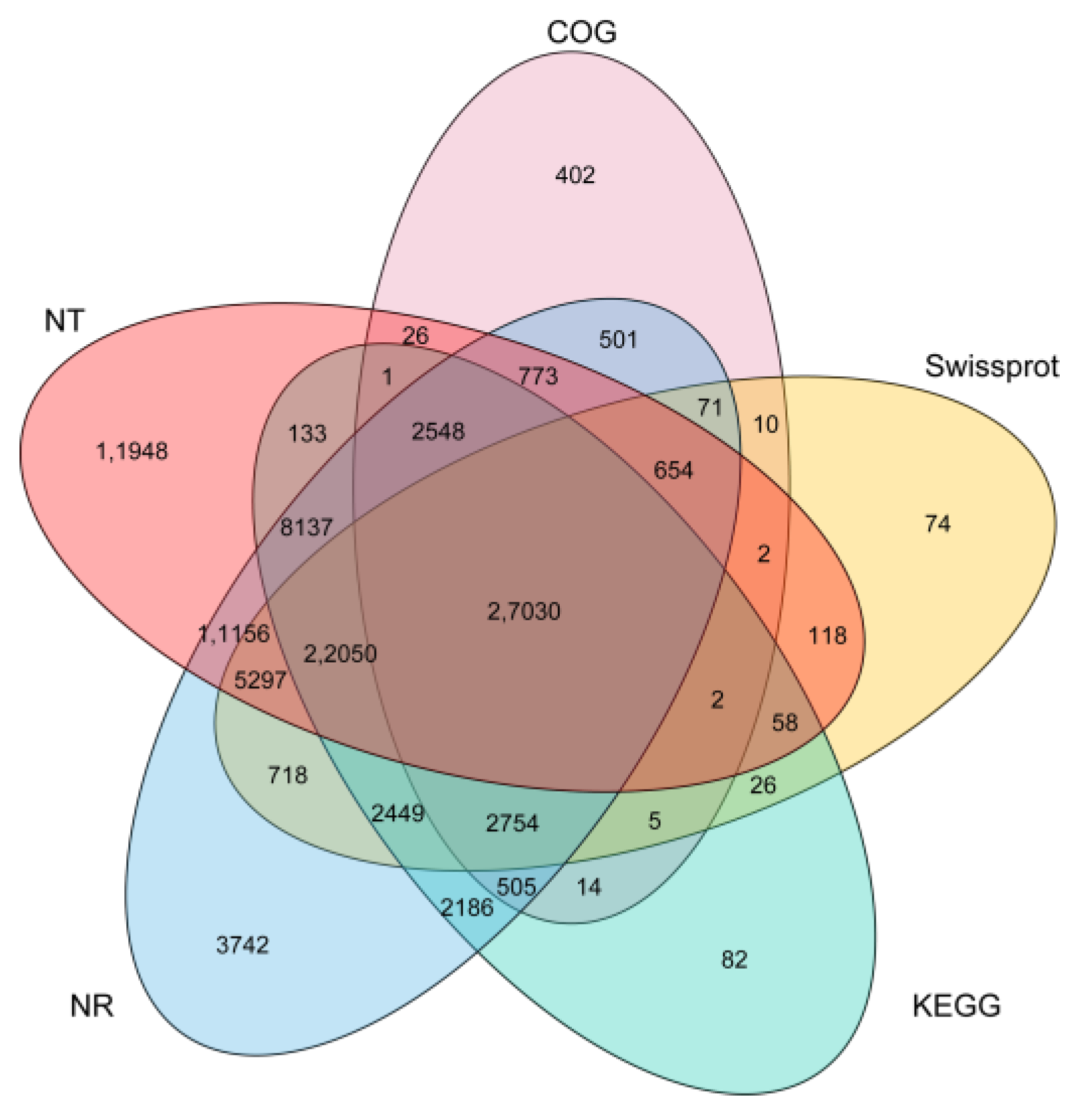
2.3. Functional Annotation
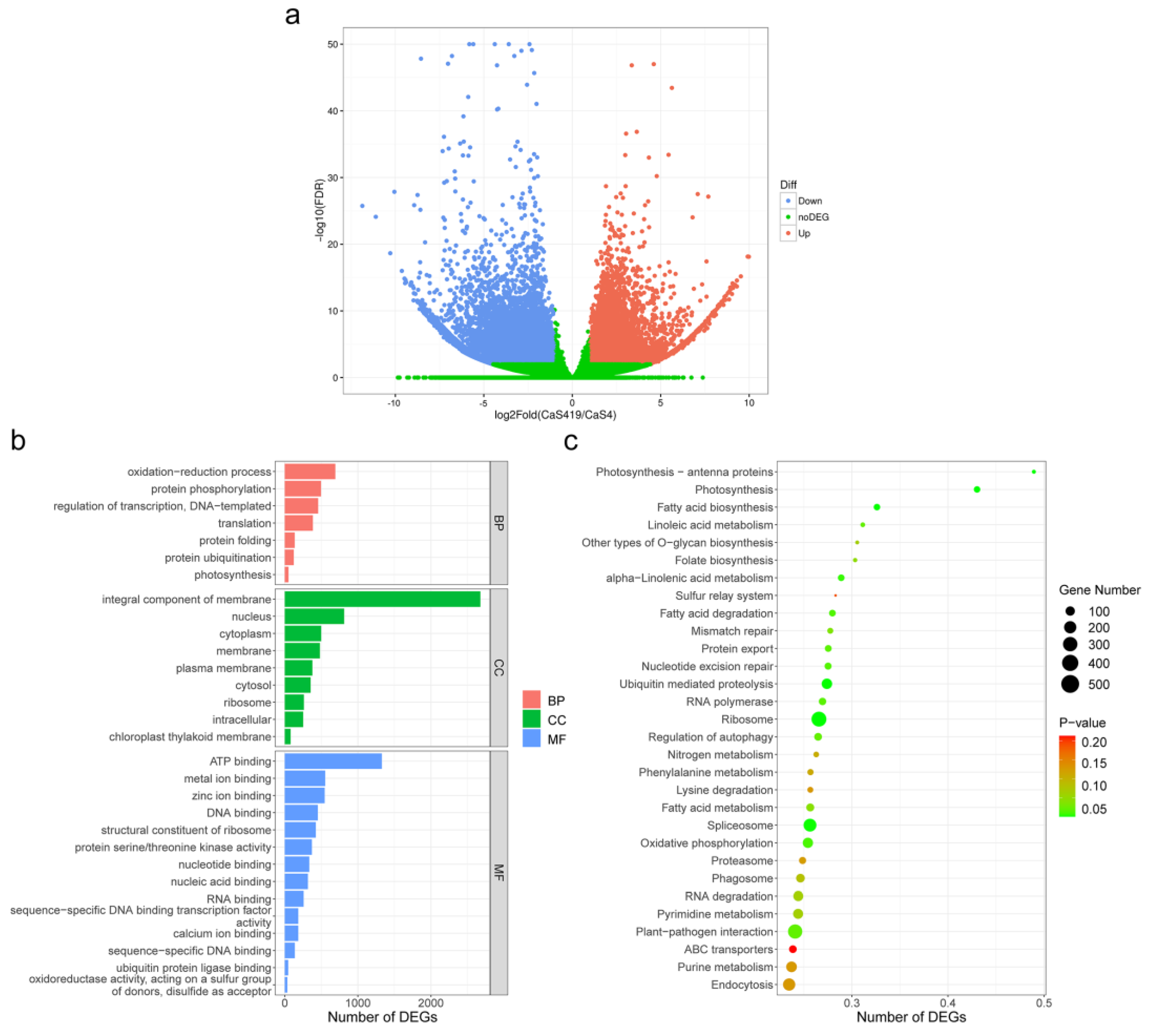
2.4. Differentially Expressed Genes (DEG) and Enrichment Analysis
2.5. DEG of Transporters Related to Cadmium (Cd), Copper (Cu) and Aluminum (Al) in Tea
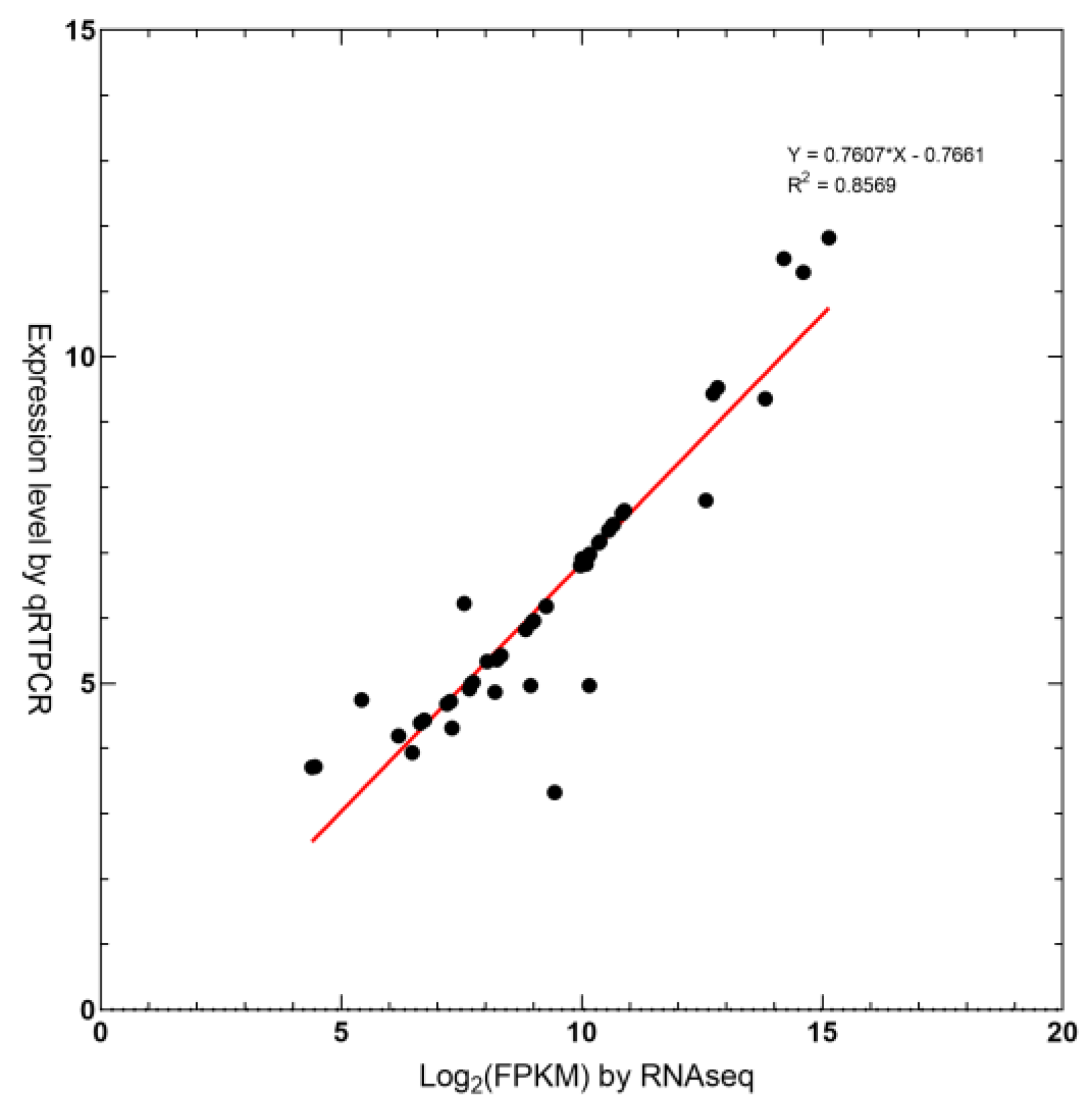
2.6. Real-Time Quantitative PCR Validation
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and Sequencing
4.3. Functional Annotation and Enrichment Analyses
4.4. Determination of Al, Cu and Cd Content in Root and Leaf
4.5. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Luo, M.; Zhu, Y.; He, Y.; Wang, Q.; Zhang, C. Transcriptome sequencing and differential gene expression analysis in Viola yedoensis Makino (Fam. Violaceae) responsive to cadmium (Cd) pollution. Biochem. Biophys. Res. Commun. 2015, 459, 60–65. [Google Scholar] [CrossRef]
- Su, Z.; Wang, X.; Yang, H.; Sun, H.; Wei, W. Transcriptome Analysis of Cadmium Exposed Jatropha curcas. China Biotechnol. 2015, 36, 69–77. [Google Scholar]
- Li, Y.; Huang, J.; Song, X.; Zhang, Z.; Jiang, Y.; Zhu, Y.; Zhao, H.; Ni, D. An RNA-Seq transcriptome analysis revealing novel insights into aluminum tolerance and accumulation in tea plant. Planta 2017, 246, 91–103. [Google Scholar] [CrossRef]
- Aydinalp, C.; Marinova, S. The effects of heavy metals on seed germination and plant growth on alfalfa plant (Medicago sativa). Bulg. J. Agric. Sci. 2009, 15, 347–350. [Google Scholar]
- Souza, V.L.; de Almeida, A.-A.F.; Lima, S.G.C.; de M. Cascardo, J.C.; da C. Silva, D.; Mangabeira, P.A.O.; Gomes, F.P. Morphophysiological Responses and Programmed Cell Death Induced by Cadmium in Genipa Americana L. (Rubiaceae). BioMetals 2011, 24, 59–71. [Google Scholar] [CrossRef]
- Perfus-Barbeoch, L.; Leonhardt, N.; Vavasseur, A.; Forestier, C. Heavy Metal Toxicity: Cadmium Permeates through Calcium Channels and Disturbs the Plant Water Status. Plant J. 2002, 32, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Ashraf, U.; Khan, I.; Wang, L. Alteration in Growth, Leaf Gas Exchange, and Photosynthetic Pigments of Maize Plants Under Combined Cadmium and Arsenic Stress. Water Air Soil Pollut. 2017, 228, 13. [Google Scholar] [CrossRef]
- Khaliq, M.A.; James, B.; Chen, Y.H.; Ahmed Saqib, H.S.; Li, H.H.; Jayasuriya, P.; Guo, W. Uptake, Translocation, and Accumulation of Cd and Its Interaction with Mineral Nutrients (Fe, Zn, Ni, Ca, Mg) in Upland Rice. Chemosphere 2019, 215, 916–924. [Google Scholar] [CrossRef]
- Guo, T.R.; Zhang, G.P.; Zhou, M.X.; Wu, F.B.; Chen, J.X. Inflfluence of Aluminum and Cadmium Stresses on Mineral Nutrition and Root Exudates in Two Barley Cultivars. Pedosphere 2007, 17, 505–512. [Google Scholar] [CrossRef]
- Zhang, Q.; Wen, Q.; Ma, T.; Zhu, Q.; Huang, D.; Zhu, H.; Xu, C.; Chen, H. Cadmium-Induced Iron Defificiency Is a Compromise Strategy to Reduce Cd Uptake in Rice. Environ. Exp. Bot. 2023, 206, 105155. [Google Scholar] [CrossRef]
- Tiwari, S.; Patel, A.; Pandey, N.; Raju, A.; Singh, M.; Prasad, S.M. Defificiency of Essential Elements in Crop Plants. In Sustainable Solutions for Elemental Defificiency and Excess in Crop Plants; Mishra, K., Tandon, P.K., Srivastava, S., Eds.; Springer: Singapore, 2020; pp. 19–52. ISBN 9789811586354. [Google Scholar]
- Jiang, X.J.; Luo, Y.M.; Liu, Q.; Liu, S.L.; Zhao, Q.G. Effects of Cadmium on Nutrient Uptake and Translocation by Indian Mustard. Environ. Geochem. Health 2004, 26, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Flora, S.J.S. Arsenic: Chemistry, Occurrence, and Exposure. In Handbook of Arsenic Toxicology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–49. ISBN 9780124186880. [Google Scholar]
- Garg, N.; Singla, P. Arsenic Toxicity in Crop Plants: Physiological Effects and Tolerance Mechanisms. Environ. Chem. Lett. 2011, 9, 303–321. [Google Scholar] [CrossRef]
- Stoeva, N.; Berova, M.; Zlatev, Z. Effect of Arsenic on Some Physiological Parameters in Bean Plants. Biol. Plant. 2005, 49, 293–296. [Google Scholar] [CrossRef]
- Nutt, L.K.; Gogvadze, V.; Uthaisang, W.; Mirnikjoo, B.; McConkey, D.J.; Orrenius, S. Bax and Bak Are Required for Cytochrome c Release during Arsenic Trioxide-Induced Apoptosis. Cancer Biol. Ther. 2005, 4, 465–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeinvand-Lorestani, M.; Kalantari, H.; Khodayar, M.J.; Teimoori, A.; Saki, N.; Ahangarpour, A.; Rahim, F.; Khorsandi, L. Dysregulation of Sqstm1, Mitophagy, and Apoptotic Genes in Chronic Exposure to Arsenic and High-Fat Diet (HFD). Environ. Sci. Pollut. Res. 2018, 25, 34351–34359. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Qi, Y.; Chen, H.; Zhang, B.; Chen, Z.; Lu, L. Study of Camellia sinensis diploid and triploid leaf development mechanism based on transcriptome and leaf characteristics. PLoS ONE 2023, 18, e0275652. [Google Scholar] [CrossRef]
- Yao, X.; Qi, Y.; Chen, H.; Zhang, B.; Chen, Z.; Lu, L. Comparative transcriptomic and proteomic analysis of nutritional quality-related molecular mechanisms of ‘Qianmei 419’ and ‘Qianfu 4’ varieties of Camellia sinensis. 2023, gene.865. Gene 2023, 865, 147329. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, H.; Wang, J. Breeding of Qianfu 4, new triploid tea variety. Southwest China J. Agric. Sci. 2009, 22, 1194–1197. [Google Scholar]
- Ali, I.; Aboul-Enein, H.Y.; Gupta, V.K. Nanochromatography and Nanocapillary Electrophoresis: Pharmaceutical and Environmental Analyses; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 978-0-470-17851-5. [Google Scholar]
- Ali, I.; Aboul-Enein, H.Y. Instrumental Methods in Metal Ion Speciation; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Dağhan, H.; Öztürk, M.; Hakeem, K.R.; Sabir, M.; Mermut, A.R. Soil pollution in Turkey and remediation methods. In Soil Remediation and Plants: Prospects and Challenges; Elsevier: Amsterdam, The Netherlands, 2015; pp. 287–312. [Google Scholar]
- Gupta, V.K.; Ali, I. Environmental Water: Advances in Treatment, Remediation and Recycling; Newnes: Oxford, UK, 2013. [Google Scholar]
- Hakeem, K.; Sabir, M.; Ozturk, M.; Mermut, A.R. Soil Remediation and Plants: Prospects and Challenges; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Hubbard, A.T. Encyclopedia of Surface and Colloid Science; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Öztürk, M.; Ashraf, M.; Aksoy, A.; Ahmad, M.S.A. Phytoremediation for Green Energy; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Alharbi, O.M.; Khattab, R.A.; Ali, I. Health and environmental effects of persistent organic pollutants. J. Mol. Liq. 2018, 263, 442–453. [Google Scholar] [CrossRef]
- Ali, I.; Alharbi, O.M.; Alothman, Z.A.; Badjah, A.Y.; Alwarthan, A. Artificial neural network modelling of amido black dye sorption on iron composite nano material: Kinetics and thermodynamics studies. J. Mol. Liq. 2018, 250, 1–8. [Google Scholar] [CrossRef]
- Ashraf, M.; Ozturk, M.; Ahmad, M.S.A. Plant Adaptation and Phytoremediation; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Burakova, E.A.; Dyachkova, T.P.; Rukhov, A.V.; Tugolukov, E.N.; Galunin, E.V.; Tkachev, A.G.; Ali, I. Novel and economic method of carbon nanotubes synthesis on a nickel magnesium oxide catalyst using microwave radiation. J. Mol. Liq. 2018, 253, 340–346. [Google Scholar] [CrossRef]
- Krzesłowska, M. The cell wall in plant cell response to trace metals: Polysaccharide remodeling and its role in defense strategy. Acta Physiol. Plant. 2011, 33, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Öztürk, M.A. Plants and Pollutants in Developed and Developing Countries; Ege University, Botany Department: Izmir, Turkey, 1989. [Google Scholar]
- Sabir, M.; Waraich, E.A.; Hakeem, K.R.; Öztürk, M.; Ahmad, H.R.; Shahid, M. Phytoremediation: Mechanisms and adaptations. Soil Remediat. Plants Prospect. Chall. 2014, 85, 85–105. [Google Scholar]
- Suhail, M.; Ali, I. Advanced spiral periodic classification of the elements. Chem. Int. 2017, 3, 220–224. [Google Scholar]
- Adriano, D.C. Trace Elements in the Terrestrial Environment; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Gough, L.P.; Shacklette, H.T.; Case, A.A. Element Concentrations Toxic to Plants, Animals and Man; US Government Printing Office: Washington, DC, USA, 1979. [Google Scholar]
- Gucel, S.; Ozturk, M.; Yucel, E.; Kadis, C.; Guvensen, A. Studies on the trace metals in the soils and plants growing in the vicinity of copper mining area-Lefke, Northern Cyprus. Frese Environ. Bull. 2009, 18, 360–368. [Google Scholar]
- SHARMA, S.; Imran, A.L.I. Adsorption of Rhodamine B dye from aqueous solution onto acid activated mango (Magnifera indica) leaf powder: Equilibrium, kinetic and thermodynamic studies. J. Toxicol. Environ. Health Sci. 2011, 3, 286–297. [Google Scholar]
- Ghori, N.H.; Ghori, T.; Hayat, M.Q.; Imadi, S.R.; Gul, A.; Altay, V.; Ozturk, M. Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 2019, 16, 1807–1828. [Google Scholar] [CrossRef]
- Fu, J.; Yu, D.; Chen, X.; Su, Y.; Li, C.; Wei, Y. Recent research progress in geochemical properties and restoration of heavy metals in contaminated soil by phytoremediation. J. Mt. Sci. 2019, 16, 2079–2095. [Google Scholar] [CrossRef]
- Qiao, J.; Zhang, Y.; Zhu, X.; Yang, H.; Zeng, W.; Zhao, Q.; Yuan, H. Identification of genes up-regulated by Ectropic obliqua feeding in tea seedlings. Acta Hortic. Sin. 2011, 38, 783–789. [Google Scholar]
- Wei, C.L.; Gao, X.F.; Ye, A.H.; Yang, Y.Q.; Jiang, C.J. Differential gene expressionprofiles analysis of tea plant induced by tea looper (Ectropic oblique) attackusing DDRT-PCR. J. Tea Sci. 2017, 27, 133–140. [Google Scholar]
- Peng, C.; Zhu, X.; Hou, R.; Ge, G.; Hua, R.; Wan, X.; Cai, H. Aluminum and heavy metal accumulation in tea leaves: An interplay of environmental and plant factors and an assessment of exposure risks to consumers. J. Food Sci. 2018, 83, 1165–1172. [Google Scholar] [CrossRef]
- Cao, D.; Liu, Y.; Ma, L.; Jin, X.; Guo, G.; Tan, R.; Liu, Z.; Zheng, L.; Ye, F.; Liu, W. Transcriptome analysis of differentially expressed genes involved in selenium accumulation in tea plant (Camellia sinensis). PLoS ONE 2018, 13, e0197506. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.L.; Li, Q.H.; Xin, H.H.; Chen, X.; Zhu, X.J.; Li, X.H. Reliable reference genes for normalization of gene expression data in tea plants (Camellia sinensis) exposed to metal stresses. PLoS ONE 2017, 12, e0175863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.Y.; You, S.H.; Xu, J.; Li, Y.; He, C.J. Advances in Researching Cultivation Techniques on the Hyperaccumulators Phytoremediation of Heavy Metal. Adv. Mater. Res. 2014, 955–959, 1903–1906. [Google Scholar] [CrossRef]
- Jain, M. A next-generation approach to the characterization of a non-model plant transcriptome. Curr. Sci. 2011, 101, 1435–1439. [Google Scholar]
- Yu, X.Z.; Yao, C.; Burkard, G. Research advances on the mechanisms of heavy metal tolerance in plants. J. Integr. Plant Biol. 1999, 41, 453–457. [Google Scholar] [CrossRef]
- Zhang, F.Q.; Wang, Y.S.; Lou, Z.P.; Dong, J.D. Effect of heavy metal stress on antioxidative enzymes and lipid peroxidation in leaves and roots of two mangrove plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 2007, 67, 44–50. [Google Scholar] [CrossRef]
- Qian, H.; Li, J.; Sun, L.; Chen, W.; Sheng, G.D.; Liu, W.; Fu, Z. Combined effect of copper and cadmium on Chlorella vulgaris growth and photosynthesis-related gene transcription. Aquat. Toxicol. 2009, 94, 56–61. [Google Scholar] [CrossRef]
- Burzyński, M.; Żurek, A. Effects of copper and cadmium on photosynthesis in cucumber cotyledons. Photosynthetica 2007, 45, 239–244. [Google Scholar] [CrossRef]
- Barón, M.; Arellano, J.B.; Gorgé, J.L. Copper and photosystem II: A controversial relationship. Physiol. Plant. 1995, 94, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Xu, X.; Hu, X.; Liu, Q.; Wang, Z.; Zhang, H.; Wang, H.; Wei, M.; Wang, H.Z.; Liu, H. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa. Front. Plant Sci. 2015, 6, 1149. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.F.; Valdes, B.; Duke, M.; Peaston, K.A.; Lahner, B.; Salt, D.E.; Williams, L.E. Functional significance of AtHMA4 C-terminal domain in planta. PLoS ONE 2010, 5, e13388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, T.; Sakurai, N.; Fujita, M.; Shinozaki, K.; Shibata, D.; Kobayashi, M. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef] [Green Version]

| Assembly Feature | Value |
|---|---|
| Total_Number | 154,097 |
| Min_Length | 200 |
| Max_length | 20,083 |
| Mean_Length | 825.67 |
| N50 | 1349 |
| N90 | 332 |
| GC | 0.411 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, X.; Chen, H.; Zhang, B.; Lu, L. Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Aluminum, Copper and Cadmium Accumulation in Tea ‘Qianmei 419’ and ‘Qianfu 4’. Plants 2023, 12, 2580. https://doi.org/10.3390/plants12132580
Yao X, Chen H, Zhang B, Lu L. Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Aluminum, Copper and Cadmium Accumulation in Tea ‘Qianmei 419’ and ‘Qianfu 4’. Plants. 2023; 12(13):2580. https://doi.org/10.3390/plants12132580
Chicago/Turabian StyleYao, Xinzhuan, Hufang Chen, Baohui Zhang, and Litang Lu. 2023. "Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Aluminum, Copper and Cadmium Accumulation in Tea ‘Qianmei 419’ and ‘Qianfu 4’" Plants 12, no. 13: 2580. https://doi.org/10.3390/plants12132580
APA StyleYao, X., Chen, H., Zhang, B., & Lu, L. (2023). Transcriptome Analysis Reveals Differentially Expressed Genes Involved in Aluminum, Copper and Cadmium Accumulation in Tea ‘Qianmei 419’ and ‘Qianfu 4’. Plants, 12(13), 2580. https://doi.org/10.3390/plants12132580






