Protein–Protein Interactions and Quantitative Phosphoproteomic Analysis Reveal Potential Mitochondrial Substrates of Protein Phosphatase 2A-B’ζ Holoenzyme
Abstract
1. Introduction
2. Methodology
2.1. Plant Material and Growth Conditions
2.2. Gene Cloning for in Planta Expression In Vivo
2.3. Sugar Dependence Assay
2.4. Quantitative Phosphoproteomics
2.5. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
2.6. Bioinformatics
3. Results and Discussion
3.1. Sugar Dependence Assay Points to a Role in Energy Metabolism
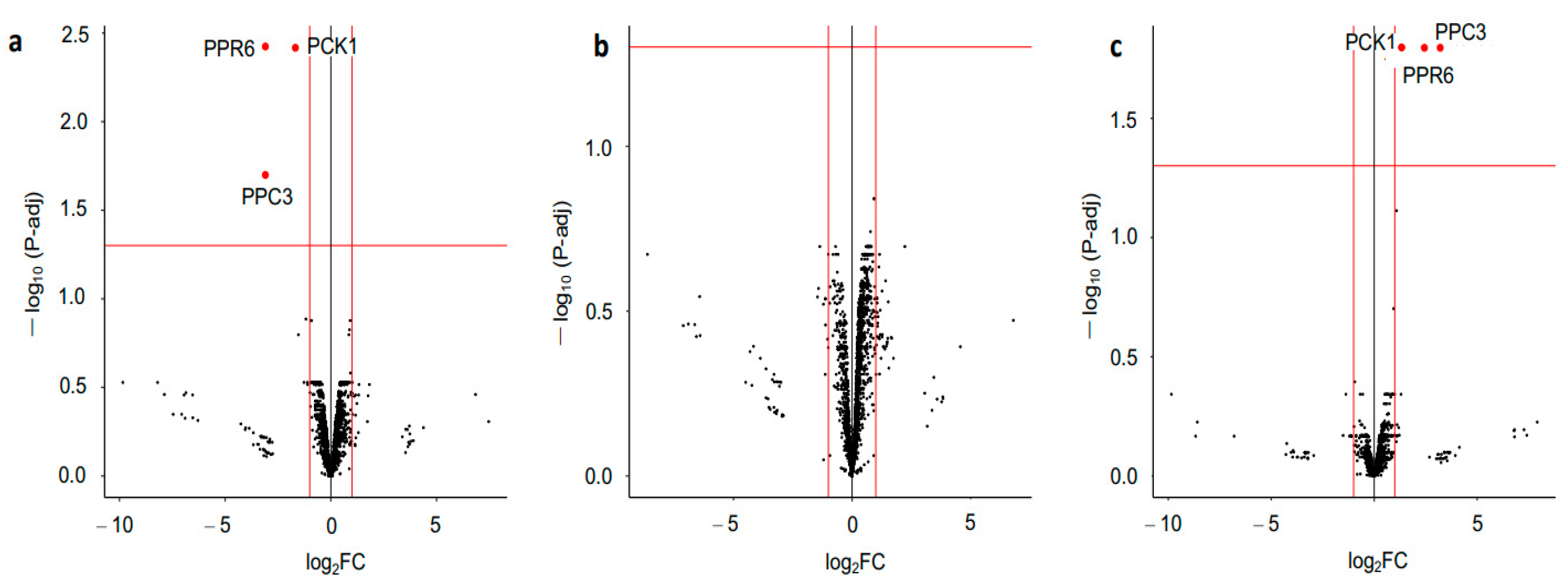
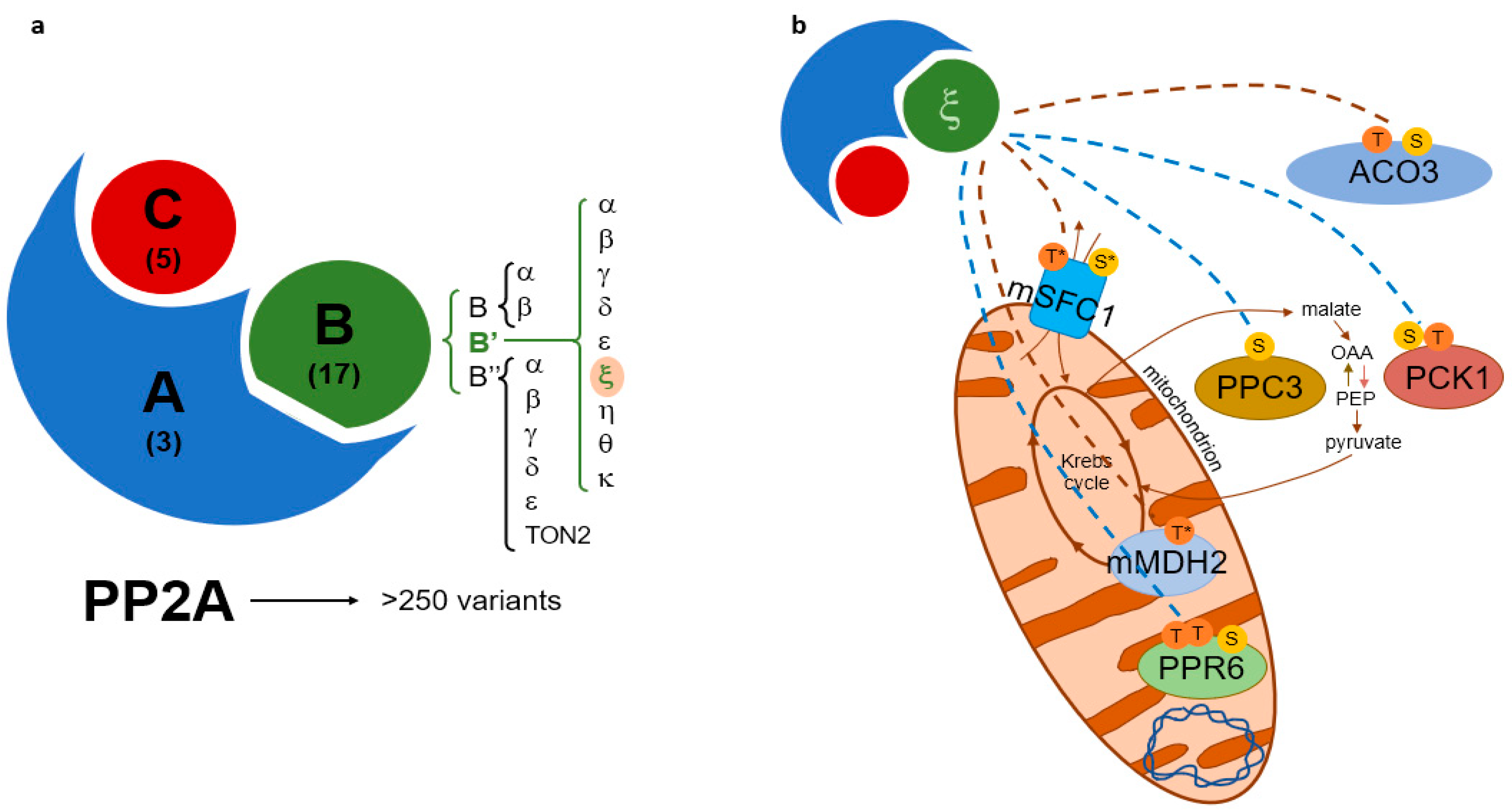
3.2. Selected B’ζ Putative Interactors Carry Ser and Thr Phosphorylation Sites
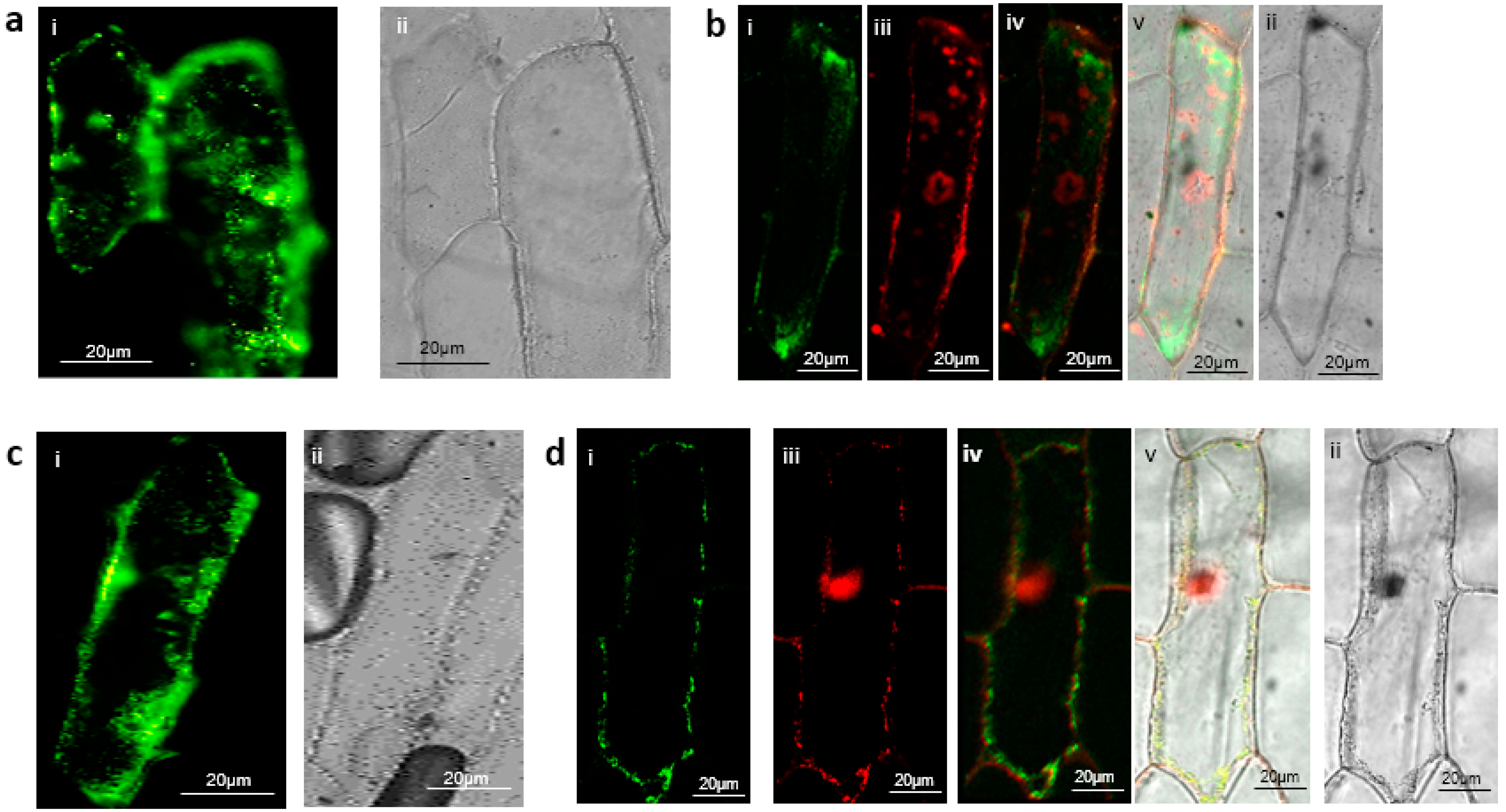
3.3. Verification of B’ζ Putative Interactors Involved in Energy Flow to Mitochondria
3.4. Additional Metabolic Enzymes Revealed by Phosphoproteomics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kinoshita-Kikuta, E.; Kinoshita, E.; Koike, T. Phospho-Proteomics Methods and Protocols, 2nd ed.; von Stechow, L., Ed.; Springer: New York, NY, USA, 2016; ISBN 978-1-4939-3048-7. [Google Scholar]
- Bheri, M.; Mahiwal, S.; Sanyal, S.K.; Pandey, G.K. Plant protein phosphatases: What do we know about their mechanism of action? FEBS J. 2021, 288, 756–785. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, J. Protein phosphorylation in plant cell signaling. Methods Mol. Biol. 2021, 2358, 45–71. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 2014, 26, 2367–2389. [Google Scholar] [CrossRef]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Farkas, I.; Dombrádi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Matre, P.; Meyer, C.; Lillo, C. Diversity in subcellular targeting of the PP2A B’η subfamily members. Planta 2009, 230, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Segonzac, C.; Macho, A.P.; Sanmartín, M.; Ntoukakis, V.; Sánchez-Serrano, J.J.; Zipfel, C. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014, 33, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Kataya, A.R.; Heidari, B.; Lillo, C. Protein phosphatase 2A regulatory subunits affecting plant innate immunity, energy metabolism, and flowering time--joint functions among B’η subfamily members. Plant Signal. Behav. 2015, 10, e1026024. [Google Scholar] [CrossRef]
- Rasool, B.; Karpinska, B.; Konert, G.; Durian, G.; Denessiouk, K.; Kangasjärvi, S.; Foyer, C.H. Effects of light and the regulatory B-subunit composition of protein phosphatase 2A on the susceptibility of Arabidopsis thaliana to aphid (Myzus persicae) infestation. Front. Plant Sci. 2014, 5, 405. [Google Scholar] [CrossRef]
- Trotta, A.; Wrzaczek, M.; Scharte, J.; Tikkanen, M.; Konert, G.; Rahikainen, M.; Holmström, M.; Hiltunen, H.M.; Rips, S.; Sipari, N.; et al. Regulatory subunit B’γ of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol. 2011, 156, 1464–1480. [Google Scholar] [CrossRef]
- Rahikainen, M.; Trotta, A.; Alegre, S.; Pascual, J.; Vuorinen, K.; Overmyer, K.; Moffatt, B.; Ravanel, S.; Glawischnig, E.; Kangasjärvi, S. PP2A-B’γ modulates foliar trans-methylation capacity and the formation of 4-methoxy-indol-3-yl-methyl glucosinolate in Arabidopsis leaves. Plant J. 2017, 89, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Smith, K.R.; Lim, S.T.; Tian, R.; Lu, J.; Tan, M. Regulation of mitochondrial functions by protein phosphorylation and dephosphorylation. Cell Biosci. 2016, 6, 25. [Google Scholar] [CrossRef]
- Kataya, A.R.; Heidari, B.; Hagen, L.; Kommedal, R.; Slupphaug, G.; Lillo, C. Protein phosphatase 2A holoenzyme is targeted to peroxisomes by piggybacking and positively affects peroxisomal β-oxidation. Plant Physiol. 2015, 167, 493–506. [Google Scholar] [CrossRef]
- Kataya, A.R.A.; Elshobaky, A.; Heidari, B.; Dugassa, N.F.; Thelen, J.J.; Lillo, C. Multi-targeted trehalose-6-phosphate phosphatase I harbors a novel peroxisomal targeting signal 1 and is essential for flowering and development. Planta 2020, 251, 98. [Google Scholar] [CrossRef]
- Schweiger, R.; Schwenkert, S. Protein-protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J. Vis. Exp. 2014, 85, 51327. [Google Scholar] [CrossRef]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteom. 2015, 14, 556–571. [Google Scholar] [CrossRef]
- Zhu, Y.; Orre, L.M.; Zhou Tran, Y.; Mermelekas, G.; Johansson, H.J.; Malyutina, A.; Anders, S.; Lehtiö, J. DEqMS: A method for accurate variance estimation in differential protein expression analysis. Mol. Cell. Proteom. 2020, 19, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef]
- Heazlewood, J.L.; Durek, P.; Hummel, J.; Selbig, J.; Weckwerth, W.; Walther, D.; Schulze, W.X. PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant specific phosphorylation site predictor. Nucleic Acids Res. 2008, 36, D1015–D1021. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R Package Version 2.52.0. Available online: https://bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 7 May 2023).
- Carlson, M. org.At.tair.db: Genome Wide Annotation for Arabidopsis. R Package Version 3.8.2. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.At.tair.db.html (accessed on 7 May 2023).
- de Bellis, L.; Luvisi, A.; Alpi, A. Aconitase: To be or not to be inside plant glyoxysomes, that is the question. Biology 2020, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Pracharoenwattana, I.; Zhou, W.; Keech, O.; Francisco, P.B.; Udomchalothorn, T.; Tschoep, H.; Stitt, M.; Gibon, Y.; Smith, S.M. Arabidopsis has a cytosolic fumarase required for the massive allocation of photosynthate into fumaric acid and for rapid plant growth on high nitrogen. Plant J. 2010, 62, 785–795. [Google Scholar] [CrossRef]
- Sew, Y.S.; Ströher, E.; Fenske, R.; Millar, A.H. Loss of mitochondrial malate dehydrogenase activity alters seed metabolism impairing seed maturation and post-germination growth in Arabidopsis. Plant Physiol. 2016, 171, 849–863. [Google Scholar] [CrossRef]
- Boutry, M.; Nagy, F.; Poulsen, C.; Aoyagi, K.; Chua, N.H. Targeting of bacterial chloramphenicol acetyltransferase to mitochondria in transgenic plants. Nature 1987, 328, 340–342. [Google Scholar] [CrossRef]
- Ford, H.C.; Allen, W.J.; Pereira, G.C.; Liu, X.; Dillingham, M.S.; Collinson, I. Towards a molecular mechanism underlying mitochondrial protein import through the TOM and TIM23 complexes. eLife 2022, 11, e75426. [Google Scholar] [CrossRef]
- Brito, D.S.; Agrimi, G.; Charton, L.; Brilhaus, D.; Bitetto, M.G.; Lana-Costa, J.; Messina, E.; Nascimento, C.P.; Feitosa-Araújo, E.; Pires, M.V.; et al. Biochemical and functional characterization of a mitochondrial citrate carrier in Arabidopsis thaliana. Biochem. J. 2020, 477, 1759–1777. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, O.V.; Piroddi, M.; Galli, F.; Lushchak, V.I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep. 2014, 19, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Ben-Menachem, R.; Regev-Rudzki, N.; Pines, O. The aconitase C-terminal domain is an independent dual targeting element. J. Mol. Biol. 2011, 409, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Konert, G.; Trotta, A.; Kouvonen, P.; Rahikainen, M.; Durian, G.; Blokhina, O.; Fagerstedt, K.; Muth, D.; Corthals, G.L.; Kangasjärvi, S. Protein phosphatase 2A (PP2A) regulatory subunit B’γ interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol. 2015, 205, 1250–1263. [Google Scholar] [CrossRef] [PubMed]
- Catoni, E.; Schwab, R.; Hilpert, M.; Desimone, M.; Schwacke, R.; Flügge, U.I.; Schumacher, K.; Frommer, W.B. Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett. 2003, 534, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Urban, J. A review on recent trends in the phosphoproteomics workflow. From sample preparation to data analysis. Anal. Chim. Acta 2022, 1199, 338857. [Google Scholar] [CrossRef]
- Yang, J.; Kalhan, S.C.; Hanson, R.W. What is the metabolic role of phosphoenolpyruvate carboxykinase? J. Biol. Chem. 2009, 284, 27025–27029. [Google Scholar] [CrossRef]
- Walker, R.P.; Leegood, R.C. Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett. 1995, 362, 70–74. [Google Scholar] [CrossRef]
- Carter, P.J.; Nimmo, H.G.; Fewson, C.A.; Wilkins, M.B. Bryophyllum fedtschenkoi protein phosphatase type 2A can dephosphorylate phosphoenolpyruvate carboxylase. FEBS Lett. 1990, 263, 233–236. [Google Scholar] [CrossRef]
- Shane, M.W.; Fedosejevs, E.T.; Plaxton, W.C. Reciprocal control of anaplerotic phosphoenolpyruvate carboxylase by in vivo monoubiquitination and phosphorylation in developing proteoid roots of phosphate-deficient Harsh Hakea. Plant Physiol. 2013, 161, 1634–1644. [Google Scholar] [CrossRef]
- Moraes, T.F.; Plaxton, W.C. Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures. Eur. J. Biochem. 2000, 267, 4465–4476. [Google Scholar] [CrossRef]
- Gregory, A.L.; Hurley, B.A.; Tran, H.T.; Valentine, A.J.; She, Y.M.; Knowles, V.L.; Plaxton, W.C. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 2009, 420, 57–65. [Google Scholar] [CrossRef]
- Manavski, N.; Guyon, V.; Meurer, J.; Wienand, U.; Brettschneider, R. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell 2012, 24, 3087–3105. [Google Scholar] [CrossRef]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR proteins in organelle RNA metabolism and chloroplast biogenesis. Front. Plant Sci. 2021, 12, 627501. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elshobaky, A.; Lillo, C.; Hodén, K.P.; Kataya, A.R.A. Protein–Protein Interactions and Quantitative Phosphoproteomic Analysis Reveal Potential Mitochondrial Substrates of Protein Phosphatase 2A-B’ζ Holoenzyme. Plants 2023, 12, 2586. https://doi.org/10.3390/plants12132586
Elshobaky A, Lillo C, Hodén KP, Kataya ARA. Protein–Protein Interactions and Quantitative Phosphoproteomic Analysis Reveal Potential Mitochondrial Substrates of Protein Phosphatase 2A-B’ζ Holoenzyme. Plants. 2023; 12(13):2586. https://doi.org/10.3390/plants12132586
Chicago/Turabian StyleElshobaky, Ahmed, Cathrine Lillo, Kristian Persson Hodén, and Amr R. A. Kataya. 2023. "Protein–Protein Interactions and Quantitative Phosphoproteomic Analysis Reveal Potential Mitochondrial Substrates of Protein Phosphatase 2A-B’ζ Holoenzyme" Plants 12, no. 13: 2586. https://doi.org/10.3390/plants12132586
APA StyleElshobaky, A., Lillo, C., Hodén, K. P., & Kataya, A. R. A. (2023). Protein–Protein Interactions and Quantitative Phosphoproteomic Analysis Reveal Potential Mitochondrial Substrates of Protein Phosphatase 2A-B’ζ Holoenzyme. Plants, 12(13), 2586. https://doi.org/10.3390/plants12132586





