Eukaryotic Translation Elongation Factor OsEF1A Positively Regulates Drought Tolerance and Yield in Rice
Abstract
1. Introduction
2. Results
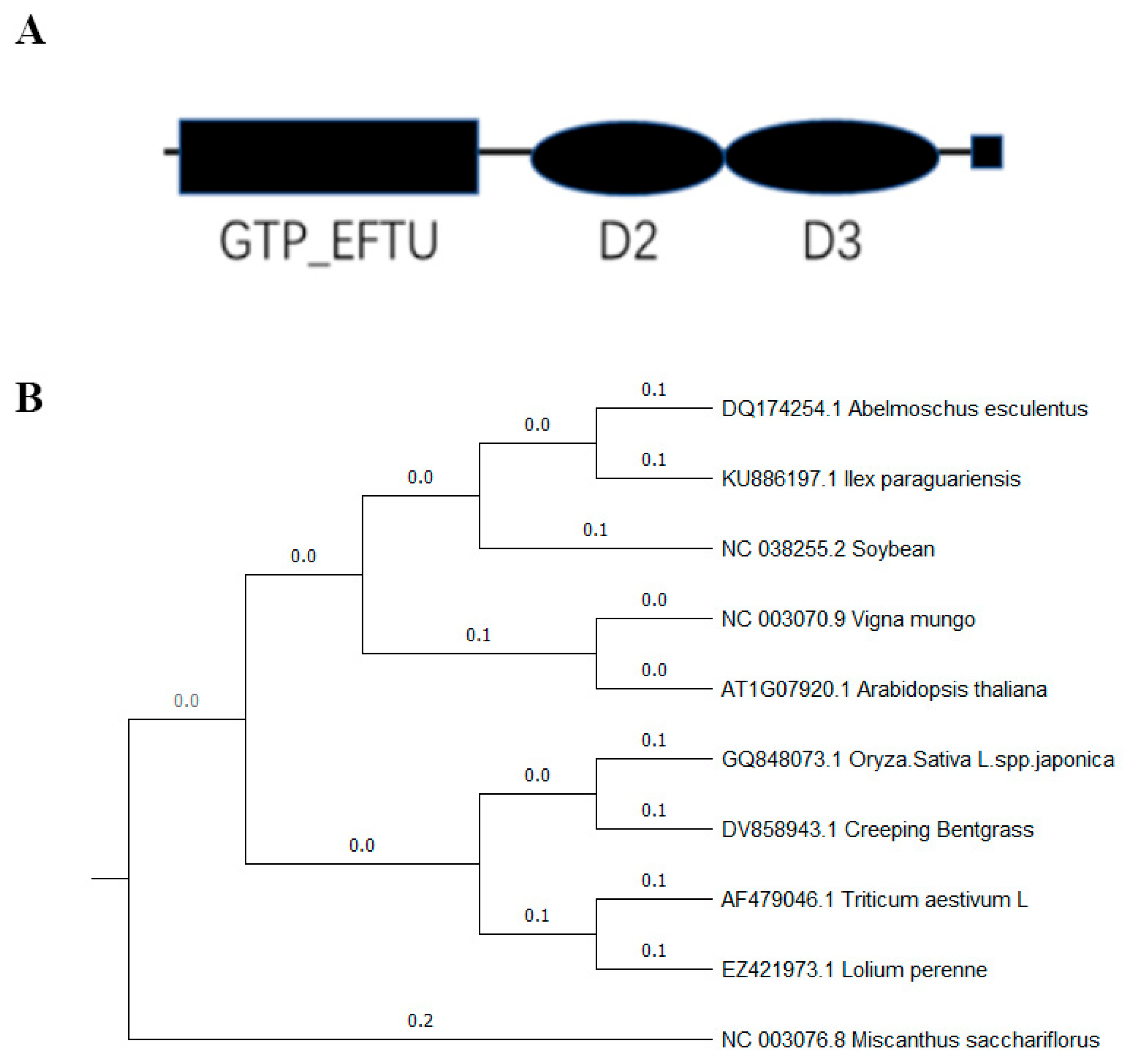
2.1. Confirmation of OsEF1A as a Translation Elongation Factor
2.2. Characteristics of OsEF1A in Rice
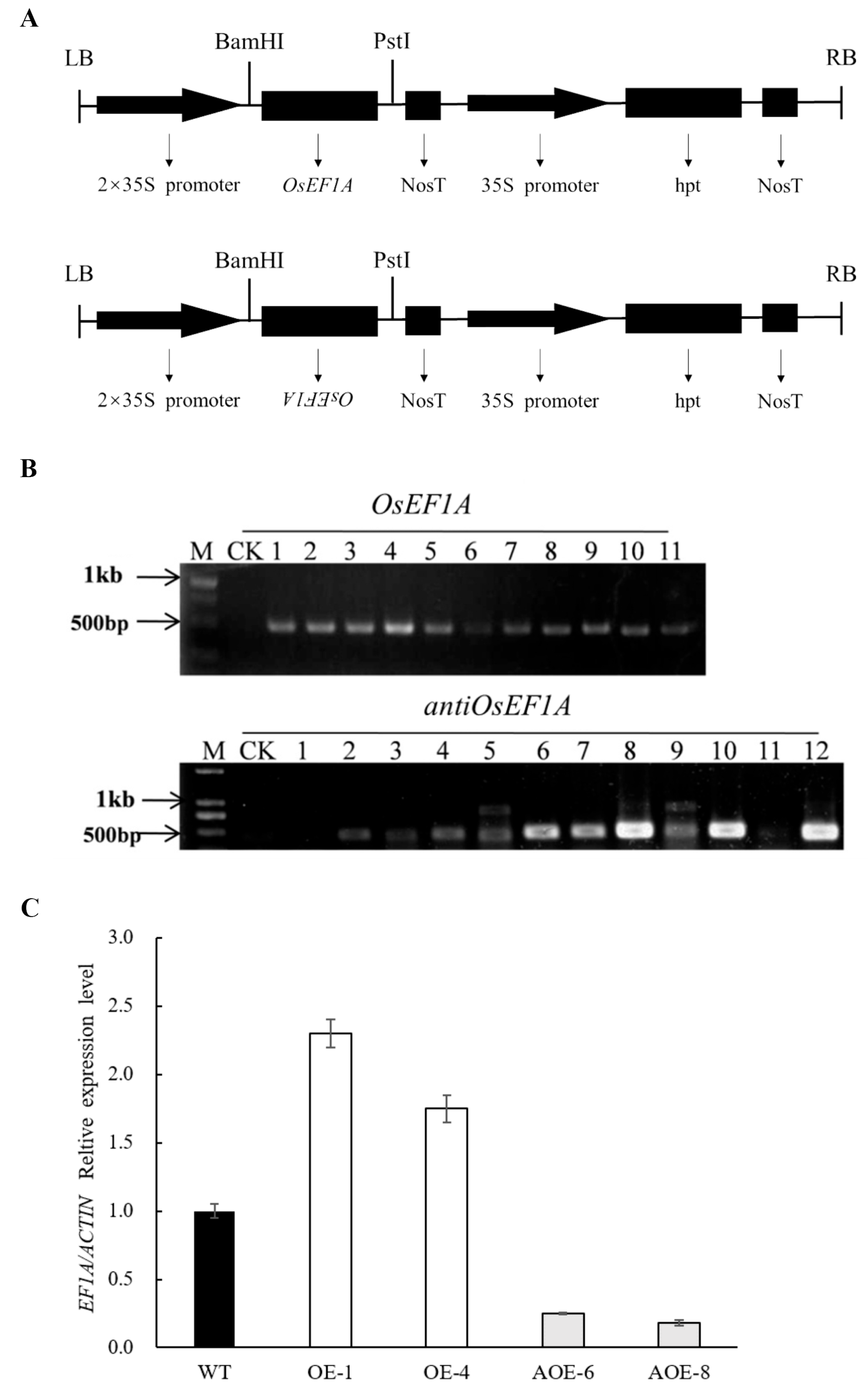
2.3. Plasmid Construction and Rice Transformation
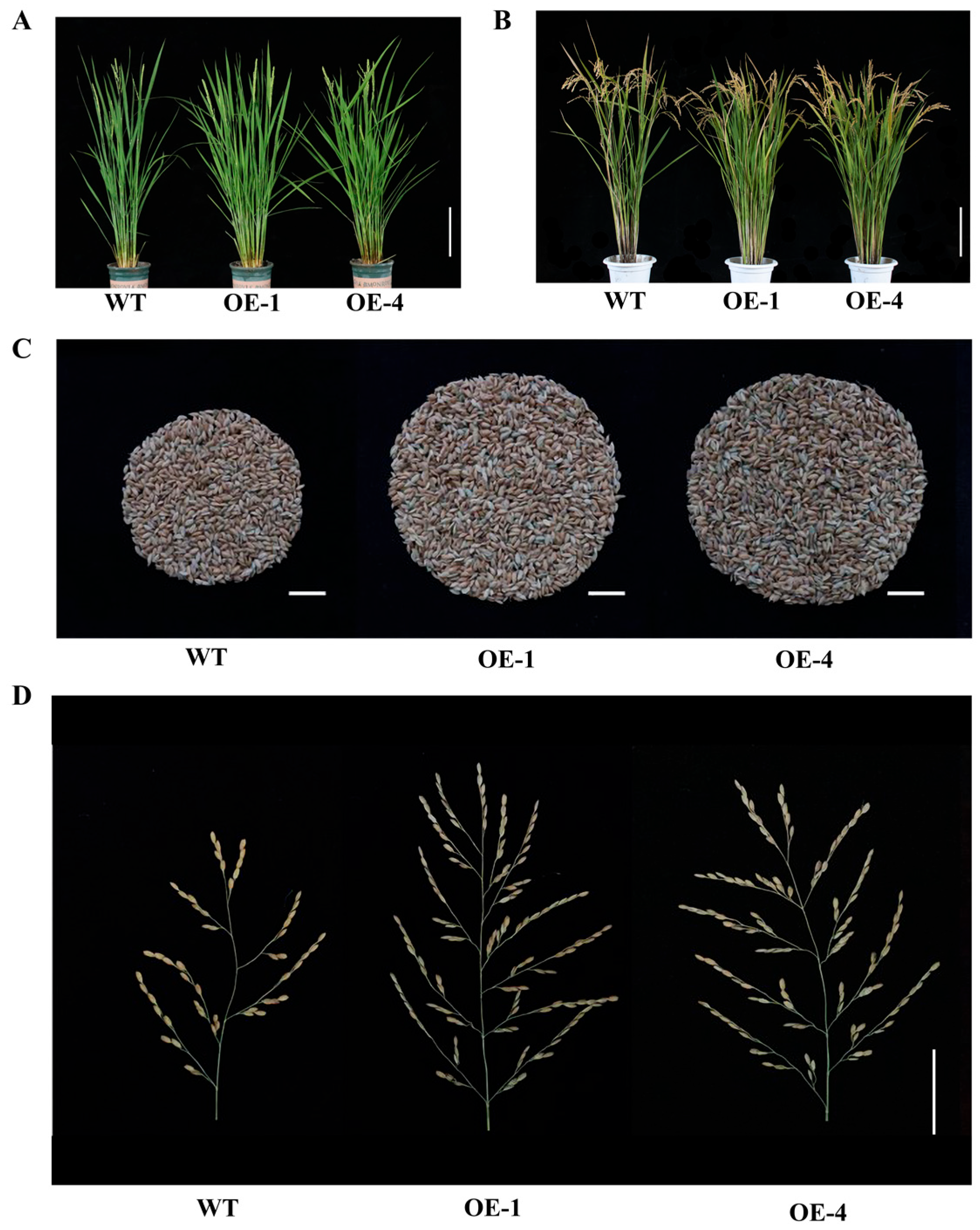
2.4. Overexpression of OsEF1A Improves Rice Yield
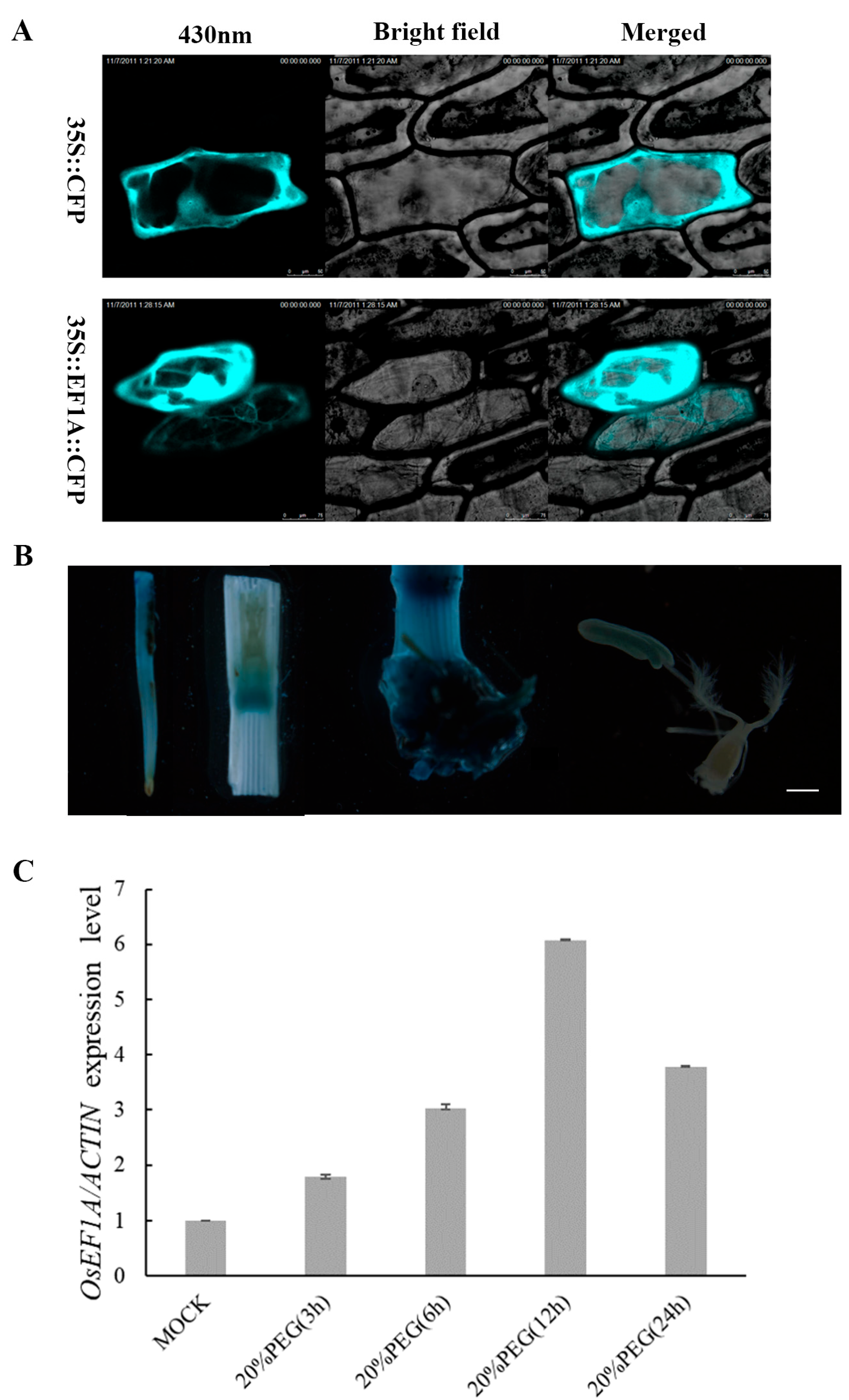
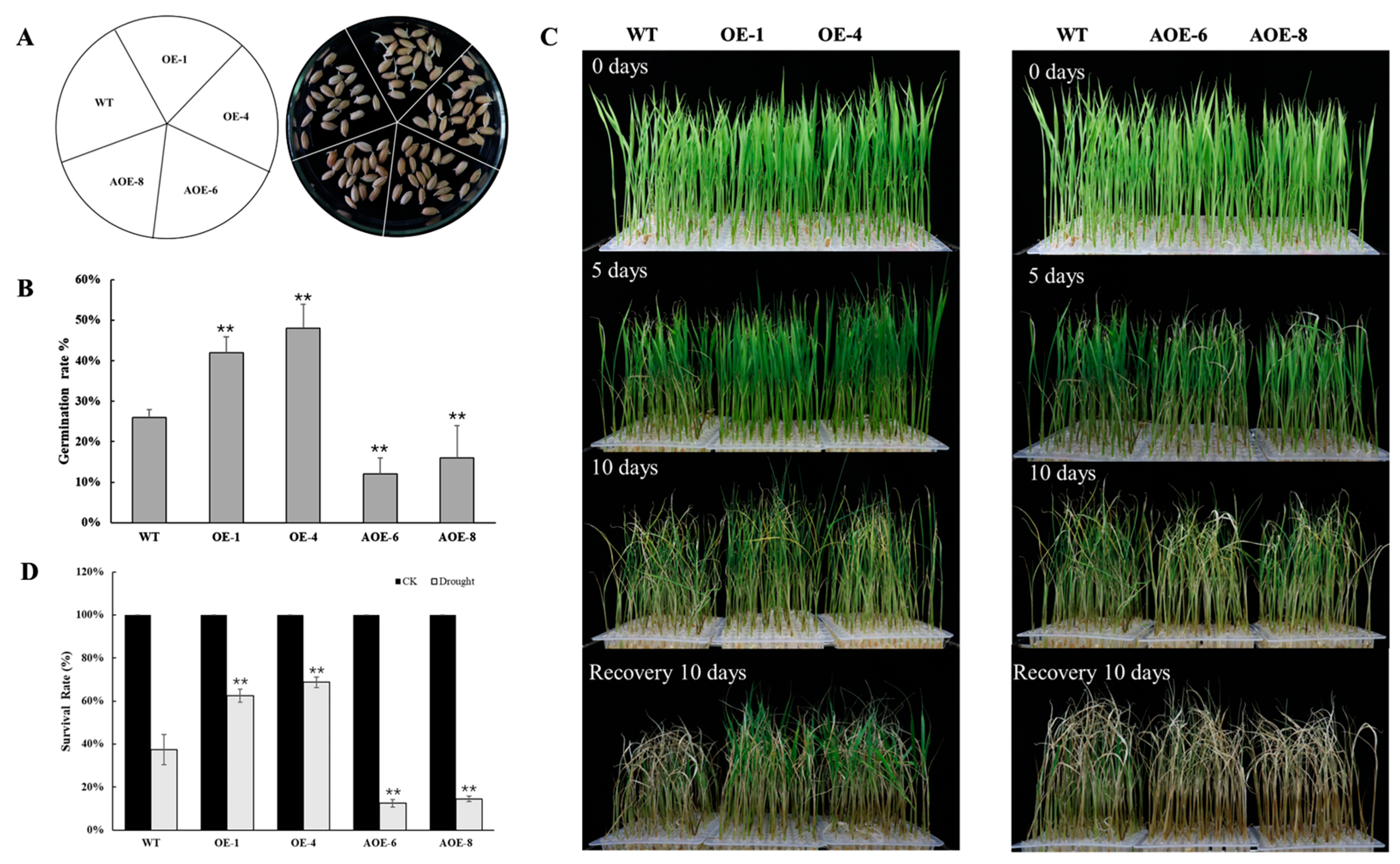
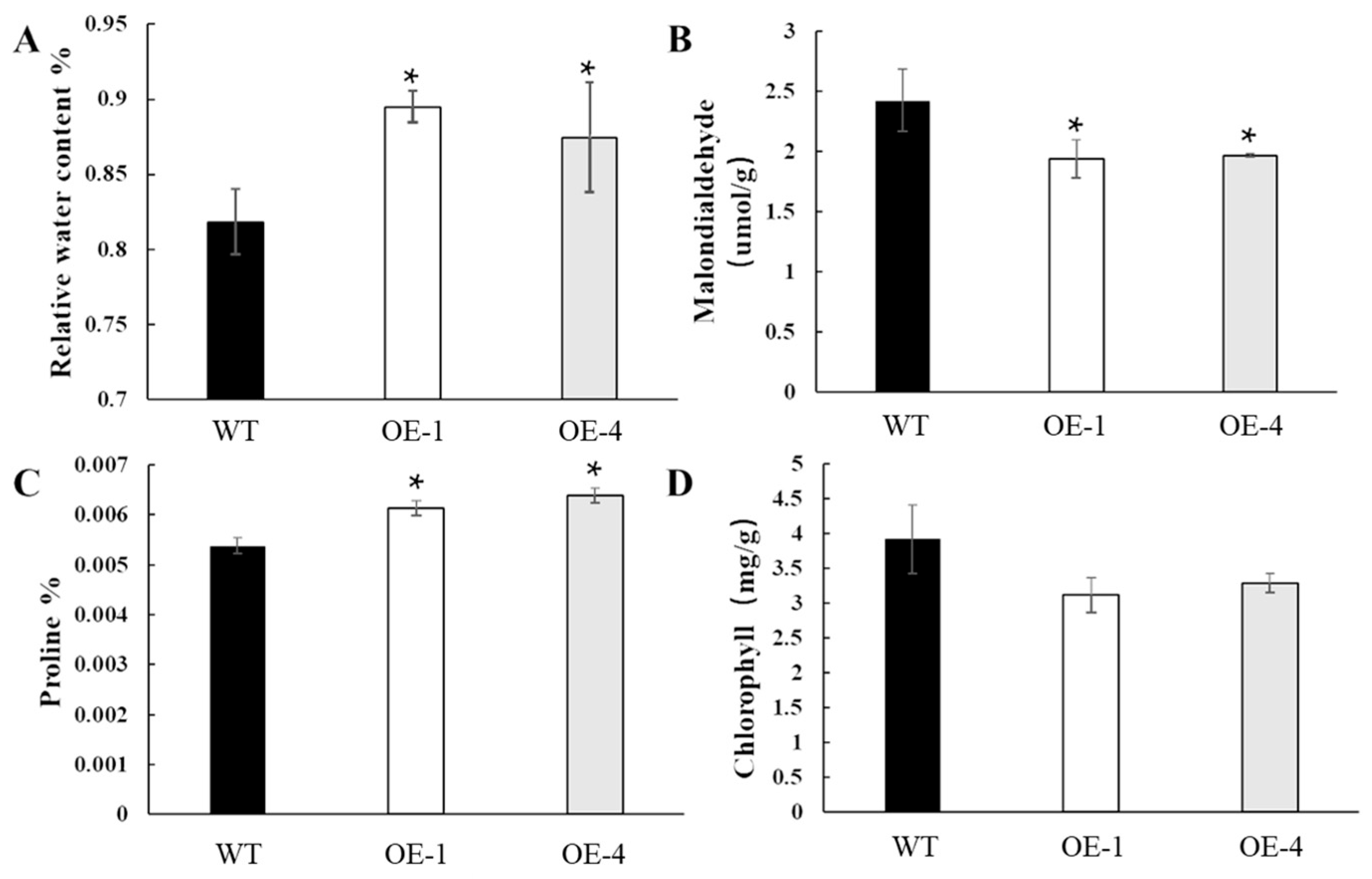
2.5. Overexpression of OsEF1A Improves Drought Resistance in Rice
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction and Transformation
4.2. Subcellular Localization of OsEF1A
4.3. Promoter–GUS Analysis
4.4. Total RNA Extraction, cDNA Synthesis and Real-Time Quantitative PCR Analysis
4.5. Growing Conditions
4.6. Drought Stress Treatment
4.7. Measurements of Agronomic Traits
4.8. Analysis of the Relative Water, Chlorophyll, Prolinecontent and Malondialdehyde Content
4.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.Y.; Lu, Z.X.; Chen, H.P. Effects of different drought stresses on root tip growth of the main root of rice seedlings. Mol. Plant Breed. 2022, 19, 6451–6458. [Google Scholar]
- Ni, S.H.; Wang, H.L.; Liu, J.N.; Gu, Y. Analysis of the characteristics and causes of agricultural drought disasters in China. Chin. Agron. Bull. 2022, 38, 106–111. [Google Scholar]
- Swamy, B.P.M.; Kumar, A. Genomics-based precision breeding approaches to improve drought tolerance in rice. Biotechnol. Adv. 2013, 31, 1308–1318. [Google Scholar] [CrossRef]
- Hu, L.L.; Li, J.; Zhang, M.S.; Zhang, H.; Yang, K. Selection and breeding of high-yielding and high-quality drought-tolerant rice variety Yongdry 1 and dry-seeding cultivation technology. Anhui Agron. Bull. 2021, 27, 87–108. [Google Scholar]
- Huang, D.Y. Water-saving and drought-resistant high-yield cultivation techniques for Shouxian Green 639 rice. Mod. Agric. Technol. 2021, 14, 15–18. [Google Scholar]
- Huang, J.P.; Wang, B.F.; Zhao, X.L.; Zhao, F.; Chen, S.Y.; Li, Y.; Cheng, J.P. Research on drought-resistant and water-saving rice cultivation and genetic breeding technology. Hubei Agric. Sci. 2015, 54, 6113–6117. [Google Scholar]
- Xu, W.; Tang, W.; Wang, C.; Ge, L.; Sun, J.; Qi, X.; He, Z.; Zhou, Y.; Chen, J.; Xu, Z.; et al. SiMYB56Confers Drought Stress Tolerance in Transgenic Rice by Regulating Lignin Biosynthesis and ABA Signaling Pathway. Front. Plant Sci. 2020, 11, 785. [Google Scholar] [CrossRef]
- Shukla, N.; Awasthi, R.P.; Rawat, L. Biochemical and physiological responses of rice (Oryza sativa L.) as influenced by Trichoderma Harzianum Under Drought Stress. Plant Physiol. Biochem. 2012, 54, 78–88. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Chen, L. Overexpression of the receptor-like kinase gene OsNRRB enhances drought-stress tolerance in rice. Euphytica 2017, 213, 86. [Google Scholar] [CrossRef]
- Suin, Y.; Dong-Keun, L.; In, J.Y.; Youn, S.K.; Yang, D.C.; Ju-Kon, K. Overexpression of the OsbZIP66 transcription factor enhances drought tolerance of rice plants. Plant Biotechnol. 2017, 11, 53–62. [Google Scholar]
- Zha, X.; Luo, X.; Qian, X.; He, G.; Yang, M.; Li, Y.; Yang, J. Over-expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 2009, 7, 611–620. [Google Scholar] [CrossRef]
- Kang, J.F.; Li, J.M.; Gao, S.; Tian, C.; Zha, X. Overexpression of the leucine-rich receptor-like kinasegene LRK2 increases drought tolerance and tiller number in rice. Plant Biotechnol. J. 2017, 15, 1175–1185. [Google Scholar] [CrossRef]
- Riis, B.; Rattan, S.I.; Clark, B.F. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990, 15, 420–424. [Google Scholar] [CrossRef]
- Gregers, R.A.; Louis, V.; Lise, P.; Kinzy, T.G.; Nyborg, J. Crystal structures of nucleotide exchange intermediates in the eEF1A–eEF1Bα complex. Nat. Struct. Biol. 2001, 8, 531–534. [Google Scholar]
- Dever, T.E.; Green, R. The Elongation, Termination, and Recycling Phases of Translation in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2012, 4, a013706. [Google Scholar] [CrossRef]
- Shopan, J.; Lv, X.; Hu, Z.; Zhang, M.; Yang, J. Eukaryotic Translation Initiation Factors Shape RNA Viruses Resistance in Plants. Hortic. Plant J. 2020, 6, 81–88. [Google Scholar] [CrossRef]
- Komoda, K.; Ishibashi, K.; Kawamura-Nagaya, K.; Ishikawa, M. Possible involvement of eEF1A in Tomato spotted wilt virus RNA synthesis. Virology 2014, 468, 81–87. [Google Scholar] [CrossRef]
- Stoianov, A.M.; Robson, D.L.; Hetherington, A.M.; Sawyez, C.G.; Borradaile, N.M. Elongation Factor 1A-1 Is a Mediator of Hepatocyte Lipotoxicity Partly through Its Canonical Function in Protein Synthesis. PLoS ONE 2015, 10, e0131269. [Google Scholar] [CrossRef]
- Wang, S.A.; Lei, C.L.; Wang, J.L.; Yang, M.; Kong, F.; Meng, Q. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. Exp. Bot. 2017, 68, 899–913. [Google Scholar] [CrossRef]
- Sun, D.; Ji, X.; Jia, Y.; Huo, D.; Si, S.; Zeng, L.; Zhang, Y.; Niu, L. LreEF1A4, a Translation Elongation Factor from Lilium regale, Is Pivotal for Cucumber Mosaic Virus and Tobacco Rattle Virus Infections and Tolerance to Salt and Drought. Mol. Sci. 2020, 21, 2083. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.; Zhang, Z.; Dai, J. Plant protein synthesis elongation factors. Plant Physiol. Newsl. 2002, 38, 406–412. [Google Scholar]
- Sun, Y.; Carneiro, N.; Clore, A.M.; Moro, G.L.; Habben, J.E.; Larkins, B.A. Characterization of maize elongation factor 1A and its relationship to protein quality in the endosperm. Plant Physiol. 1997, 115, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, N.P.; Hughes, P.A.; Larkins, B.A. The eEFIA gene family is differentially expressed in maize endosperm. Plant Mol. Biol. 1999, 41, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Momcilovic, I.; Pantelic, D.; Zdravkovic-Korac, S.; Oljaca, J.; Rudic, J.; Fu, J. Heat-induced accumulation of protein synthesis elongation factor 1A implies an important role in heat tolerance in potato. Planta 2016, 244, 671–679. [Google Scholar] [CrossRef]
- Kidou, S.; Ejiri, S. Isolation, characterization and mRNA expression of four cDNAs encoding translation elongation factor 1A from rice (Oryza sativa L.). Plant Mol. Biol. 1988, 36, 137–148. [Google Scholar] [CrossRef]
- Li, Z.Y.; Chen, S.Y. Inducible expression of the rice translation elongation factor 1A gene under environmental factor stress. J. Bot. 1999, 41, 800–806. [Google Scholar]
- Zhang, Q.; Yao, Y.B.; Li, Y.H.; Huang, J.P.; Ma, Z.G.; Wang, Z.L.; Wang, S.P.; Wang, Y.; Zhang, Y. Causes and Changes of Drought in China: Research Progress and Prospects. J. Meteorol. Res. 2020, 34, 460–481. [Google Scholar] [CrossRef]
- He, Q.; Yang, L.; Hu, W.; Zhang, J.; Xing, Y. Overexpression of an auxin receptor OsAFB6 significantly enhanced grain yield by increasing cytokinin and decreasing auxin concentrations in rice panicle. Sci. Rep. 2018, 8, 14051. [Google Scholar] [CrossRef]
- Bang, S.W.; Lee, D.K.; Jung, H.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Suh, J.W.; Kim, J.K. Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 2019, 17, 118–131. [Google Scholar] [CrossRef]
- Kumar, V.V.S.; Yadav, S.K.; Verma, R.K.; Shrivastava, S.; Ghimire, O.; Pushkar, S.; Rao, M.V.; Kumar, T.S.; Chinnusamy, V. The abscisic acid receptor OsPYL6 confers drought tolerance to indica rice through dehydration avoidance and tolerance mechanisms. J. Exp. Bot. 2021, 72, 1411–1431. [Google Scholar] [CrossRef]
- Mao, H.D.; Jian, C.; Cheng, X.X.; Chen, B.; Mei, F.; Li, F.; Zhang, Y.; Li, S.; Du, L.; Li, T.; et al. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol. J. 2021, 20, 846–861. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, X.; Zhou, D.; Ouyang, Y.; Yao, J. Overexpression of the 16-kDa alpha-amylase/trypsin inhibitor RAG2 improves grain yield and quality of rice. Plant Biotechnol. J. 2017, 15, 568–580. [Google Scholar] [CrossRef]
- Shi, W.B.; Yang, F.; Liu, J.; Gao, J.; Chen, W.T.; Jiang, D.G. Overexpression of OsbHLH120 gene improved drought resistance at seedling stage of rice. Genom. Appl. Biol. 2019, 38, 5558–5563. [Google Scholar]
- Li, Y.; Li, T.T.; He, X.R.; Zhu, Y.; Feng, Q.; Yang, X.M.; Zhou, X.H.; Li, G.B.; Ji, Y.P.; Zhao, J.H.; et al. Blocking Osa-miR1871 enhances rice resistance against Magnaporthe oryzae and yield. Plant Biotechnol. J. 2021, 20, 646–659. [Google Scholar] [CrossRef]
- Tong, Y.; Wang, C.; Zhao, L.L.; Lian, J.; Liu, X.M. The transcription factor OsbZIP5 negatively regulates drought tolerance in rice. Chin. J. Biochem. Mol. Biol. 2021, 37, 798–810. [Google Scholar]
- Gao, H.B.; Wang, W.G.; Wang, Y.H.; Liang, Y. Molecular mechanisms underlying plant architecture and its environmental plasticity in rice. Mol. Breed. 2019, 39, 167. [Google Scholar] [CrossRef]
- Zhao, D.D.; Park, J.R.; Jang, Y.H.; Kim, E.G.; Du, X.X.; Farooq, M.; Yun, B.J.; Kim, K.M. Identification of One Major QTL and a Novel Gene OsIAA17q5 Associated with Tiller Number in Rice Using QTL Analysis. Plants 2022, 11, 538. [Google Scholar] [CrossRef]
- Wu, X.M.; Li, S.F.; Hu, R.; He, R.; Jiao, R.; Mao, Y.J.; Lu, C.M.; Hu, J.; Lin, H.; Wu, R.L.; et al. Cloning and functional study of the rice tiller regulatory gene HTD3. China Rice Sci. 2021, 35, 535–542. [Google Scholar]
- Xia, T.Y.; Chen, H.Q.; Dong, S.J.; Ma, Z.; Ren, H.; Zhu, X.; Fang, X.; Chen, F. OsWUS promotes tiller bud growth by establishing weak apical dominance in rice. Plant J. 2020, 104, 1635–1647. [Google Scholar] [CrossRef]
- Liu, Y.L.; Fan, J.Y.; Chen, X.F.; Xu, S.Y.; Zhang, D.B.; Yuan, Z. Preliminary study on the role of OsAP2-4 in regulating rice tillers development. Plant Physiol. J. 2022, 58, 817–824. [Google Scholar]
- Zhu, M.; Hu, Y.J.; Tong, A.Z.; Yan, B.; Lv, Y.; Wang, S.; Ma, W.; Cui, Z.; Wang, X. LAZY1 Controls Tiller Angle and Shoot Gravitropism by Regulating the Expression of Auxin Transporters and Signaling Factors in Rice. Plant Cell Physiol. 2021, 61, 2111–2125. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Z.; Wang, W.G.; Zhang, N.; Cai, Y.; Liang, Y.; Meng, X.; Yuan, Y.; Li, J.; Wu, D.; Wang, Y. LAZY2 controls rice tiller angle through regulating starch biosynthesis in gravity-sensing cells. New Phytol. 2021, 231, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Regulation of shoot branching by auxin. Trends Plant Sci. 2003, 8, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Zou, J.H.; Zhang, S.Y.; Zaitlin, D.; Zhu, L.H. Strigolactones are a new-defined class of plant hormones which inhibit shoot branching and mediate the interaction of plant-AM fungi and plant-parasitic weeds. Sci. China Ser. C-Life Sci. 2009, 52, 693–700. [Google Scholar] [CrossRef]
- Leyser, O. The fall and rise of apical dominance. Curr. Opin. Genet. Dev. 2005, 15, 468–471. [Google Scholar] [CrossRef]
- Xie, H.; Yang, L.; Li, Z.G. The role of proline in the development of abiotic stress tolerance in plants. Biotechnol. Bull. 2011, 2, 23–27. [Google Scholar]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Wang, Y.W.; Deng, C.; Ai, P.F.; Cui, X.A.; Zhang, Z.G. ALM1, encoding a Fe-superoxide dismutase, is critical for rice chloroplast biogenesis and drought stress response. Crop J. 2021, 9, 1018–1029. [Google Scholar] [CrossRef]
- Wang, S.J.; Zhuang, K.Y.; Zhang, S.; Yang, M.; Kong, F.; Meng, Q. Overexpression of a tomato carotenoid epsilon-hydroxylase gene (SlLUT1) improved the drought tolerance of transgenic tobacco. J. Plant Physiol. 2018, 222, 103–112. [Google Scholar] [CrossRef]
- Chen, Z.K.; Xu, W.W.; Nie, J.; Khan, A.; Cao, C.G.; Li, P. Drought Stress Intensity, Duration and Its Resistance Impact on Rice (Oryza sativa L.) Seedling. Appl. Ecol. Environ. Res. 2020, 18, 469–486. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 2016, 7, 518. [Google Scholar] [CrossRef]
- An, D.; Ma, Q.; Yan, W.; Zhou, W.; Liu, G.; Zhang, P. Divergent regulation of CBF regulon on cold tolerance and plant phenotype in cassava overexpressing Arabidopsis CBF3 gene. Front. Plant Sci. 2016, 7, 1866. [Google Scholar] [CrossRef]
| Plant Line | Tiller Number per Plant (cm) | Plant Height (cm) | 1000-Grain Weight (g) | Seed Setting Rate (%) | Grain Number per Plant | Primary Branches | Secondary Branches |
|---|---|---|---|---|---|---|---|
| WT | 16.23 ± 1.75 | 80.17 ± 2.94 | 25.68 ± 1.09 | 92.5 ± 4.7 | 1052.4 ± 123.7 | 7.83 ± 1.60 | 14.67 ± 2.42 |
| OE-1 | 21.47 ± 2.25 ** | 72.42 ± 2.77 ** | 23.20 ± 1.02 ** | 85 ± 7.26 ** | 1371.4 ± 117.6 ** | 8.83 ± 1.47 * | 17.52 ± 1.78 ** |
| OE-4 | 20.32 ± 1.97 ** | 74.13 ± 2.15 ** | 23.07 ± 1.06 ** | 87 ± 6.54 ** | 1269.2 ± 101.9 ** | 8.22 ± 1.17 * | 17.03 ± 1.26 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Q.; Kang, J.; Gao, S.; Zhao, Y.; Yi, H.; Zha, X. Eukaryotic Translation Elongation Factor OsEF1A Positively Regulates Drought Tolerance and Yield in Rice. Plants 2023, 12, 2593. https://doi.org/10.3390/plants12142593
Gu Q, Kang J, Gao S, Zhao Y, Yi H, Zha X. Eukaryotic Translation Elongation Factor OsEF1A Positively Regulates Drought Tolerance and Yield in Rice. Plants. 2023; 12(14):2593. https://doi.org/10.3390/plants12142593
Chicago/Turabian StyleGu, Qing, Junfang Kang, Shuang Gao, Yarui Zhao, Huan Yi, and Xiaojun Zha. 2023. "Eukaryotic Translation Elongation Factor OsEF1A Positively Regulates Drought Tolerance and Yield in Rice" Plants 12, no. 14: 2593. https://doi.org/10.3390/plants12142593
APA StyleGu, Q., Kang, J., Gao, S., Zhao, Y., Yi, H., & Zha, X. (2023). Eukaryotic Translation Elongation Factor OsEF1A Positively Regulates Drought Tolerance and Yield in Rice. Plants, 12(14), 2593. https://doi.org/10.3390/plants12142593






