Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer
Abstract
1. Introduction
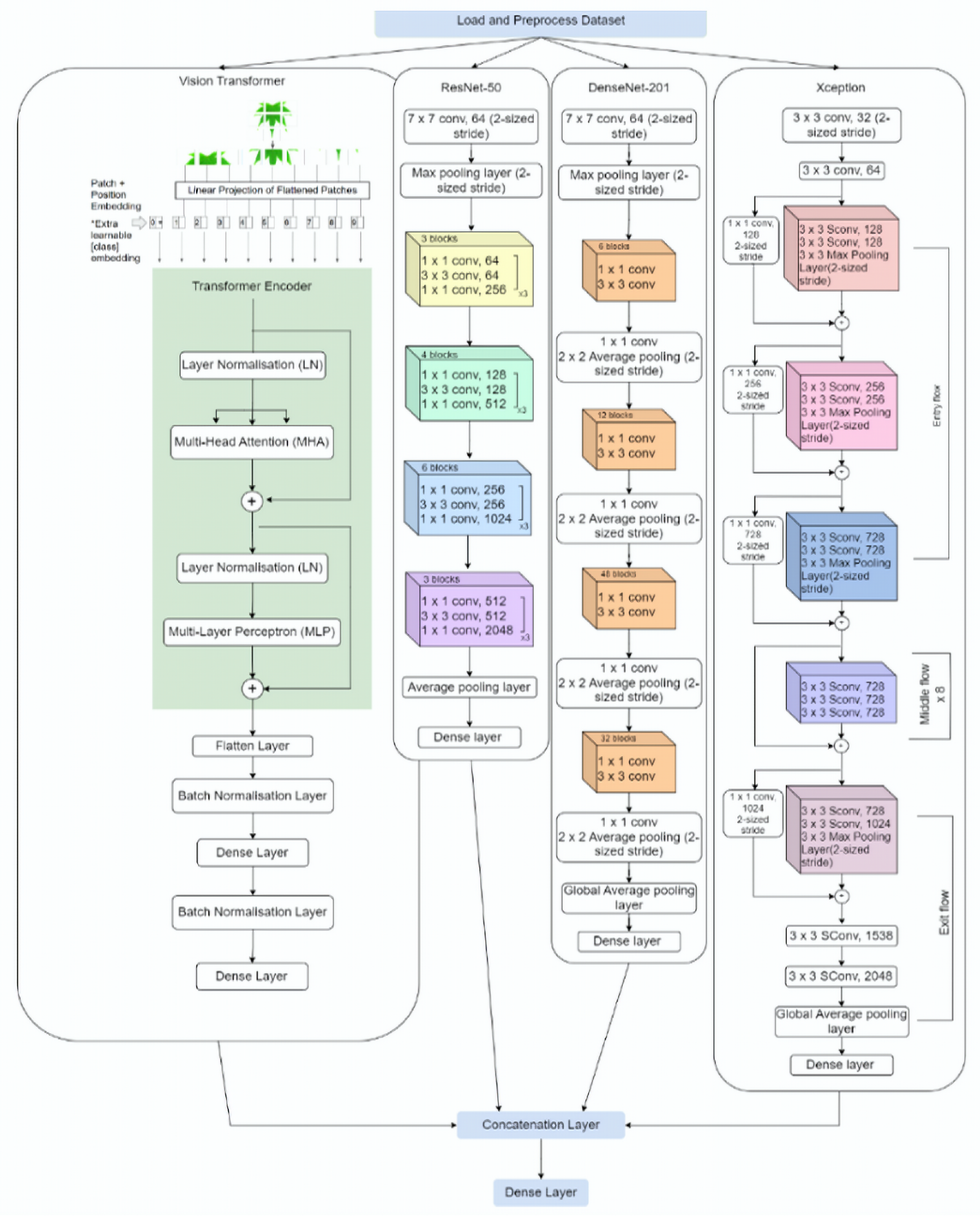
- Introduction of the Plant-CNN-ViT ensemble model, which integrates the strengths of Vision Transformer (ViT), ResNet-50, DenseNet-201, and Xception, to advance plant leaf classification. This ensemble model is designed to leverage the unique capabilities of each constituent model, enabling enhanced feature extraction and representation learning. By combining ViT, ResNet-50, DenseNet-201, and Xception, the ensemble model effectively captures the complex spatial relationships and fine-grained details present in plant leaves, leading to a notable improvement in overall classification performance.
- The efficacy of the proposed ensemble model was evaluated on widely used datasets, providing empirical evidence of its effectiveness across diverse plant species. The evaluation aimed to assess the model’s ability to handle data scarcity challenges, which are prevalent in certain datasets. Specifically, the model was subjected to the Flavia dataset, Folio Leaf dataset, Swedish Leaf dataset, and MalayaKew Leaf dataset. These datasets, with limited training samples per class, ranging from approximately 14 to 41, offer a realistic representation of the challenges encountered in plant leaf classification tasks.
2. Related Works
2.1. Traditional Machine Learning
2.2. Deep Learning
3. Plant-CNN-ViT
3.1. Vision Transformer
- Patch Embedding
- Transformer Encoder
- Classification
3.2. ResNet-50
3.3. DenseNet-201
3.4. Xception
3.5. Fusion of Model Output
4. Experiments and Discussion
4.1. Datasets
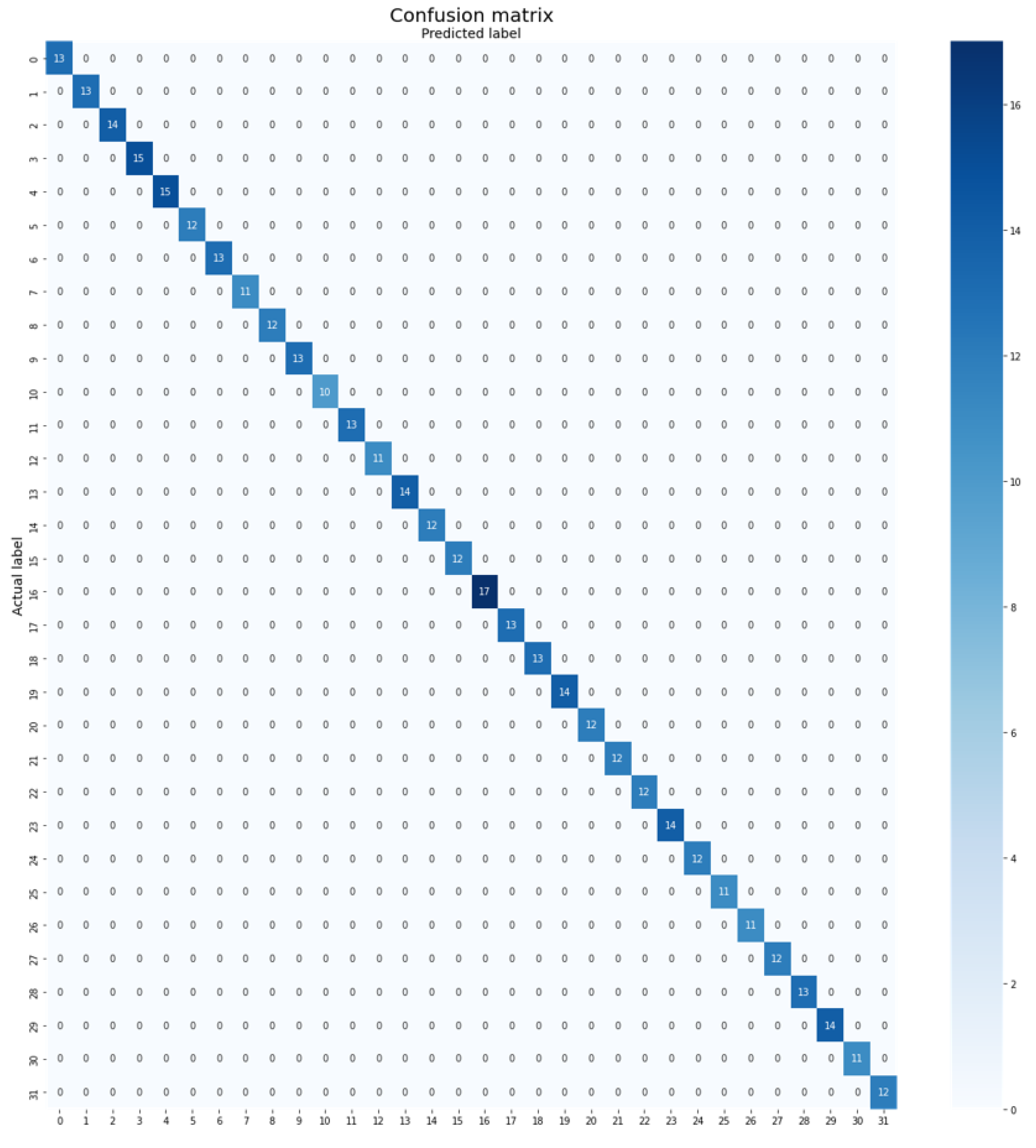
- Flavia Dataset: The Flavia dataset, created by Wu et al. [23], comprises a total of 32 classes and 1907 images. Each class contains 50 to 77 images. The dataset is divided into three subsets: training, validation, and testing. The training set consists of 1323 images (70%), the validation set contains 178 images (10%), and the testing set consists of 406 images (20%). Figure 2 presents sample images from the Flavia dataset.
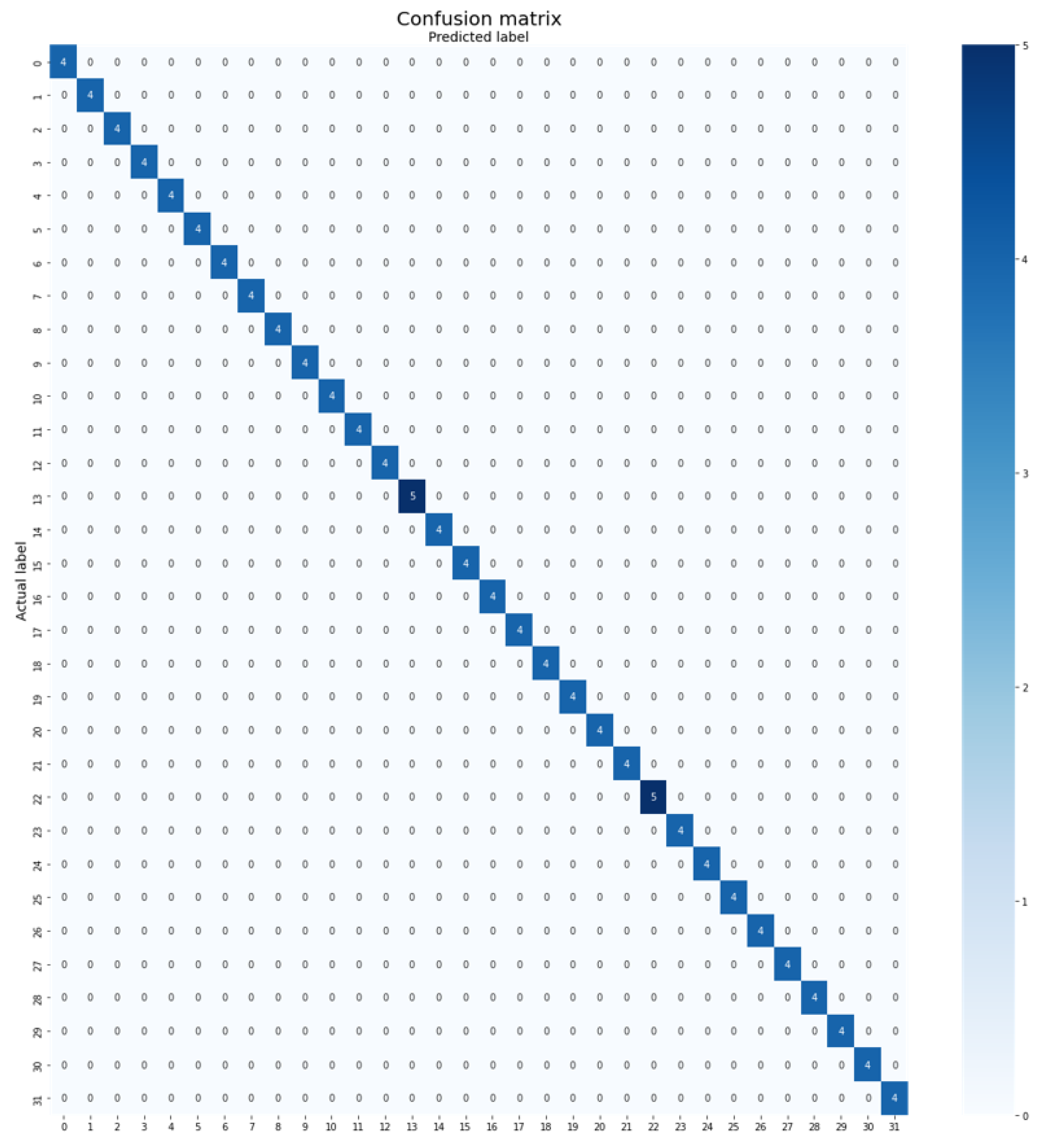
- Folio Leaf Dataset: The Folio Leaf dataset, developed by Munisami et al. [24], is the smallest dataset in this experiment, consisting of 637 images. It encompasses 32 classes, with each class containing 20 images, except for two classes. The dataset was captured in the University of Mauritius’s farm and nearby locations. The training set comprises 445 images (70%), the validation set contains 62 images (10%), and the testing set consists of 130 images (20%). Some sample images from the Folio Leaf dataset are illustrated in Figure 3.
- Swedish Leaf Dataset: The Swedish Leaf dataset, created by Oskar J. O. Söderkvist [25], includes 15 species of Swedish trees. Each class consists of 75 images, resulting in a total of 1125 images in this dataset. The dataset is divided into three sets, with 675 images in the training set (60%), 105 images in the validation set (10%), and 345 images in the testing set (30%). Figure 4 shows sample images from the Swedish Leaf dataset.
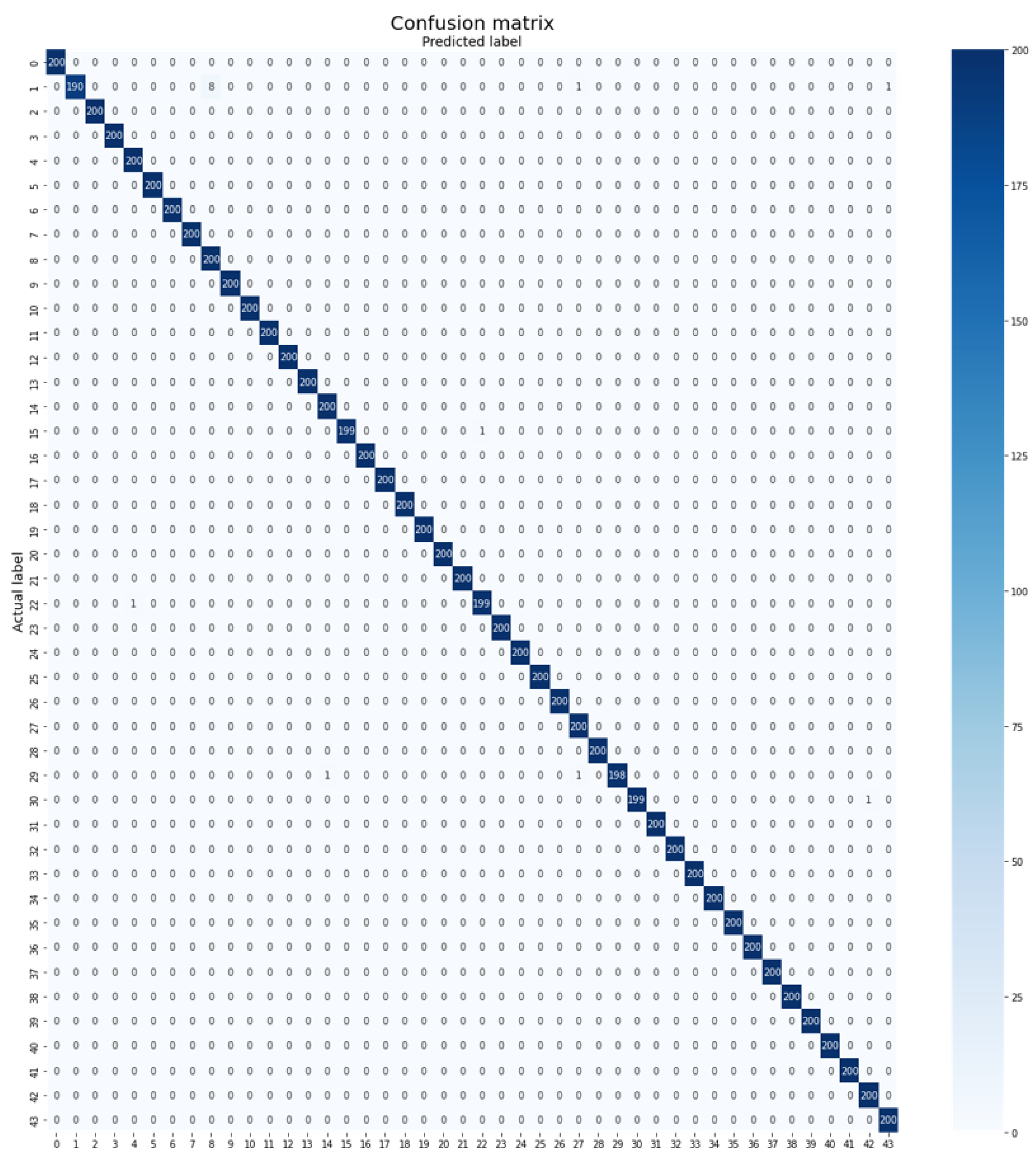
- MalayaKew Leaf Dataset: The MalayaKew Leaf dataset, introduced by Lee et al. (2015) [8], was collected from the Royal Botanical Gardens, Kew, England. It comprises two variations, namely D1 and D2, where D1 represents whole leaf images and D2 contains patches extracted from the leaves. For this experiment, D2 is employed, which includes 43,472 patches from 44 different classes. The training dataset consists of 34,672 images, while the testing dataset consists of 8800 images. The training dataset is further divided into two parts, with 80% (27,720 images) allocated to training and 20% (6952 images) for the validation set. Figure 5 showcases sample images from the MalayaKew Leaf dataset (D2).
4.2. Comparison with Existing Methods
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arafat, S.Y.; Saghir, M.I.; Ishtiaq, M.; Bashir, U. Comparison of techniques for leaf classification. In Proceedings of the 2016 Sixth International Conference on Digital Information and Communication Technology and Its Applications (DICTAP), Konya, Turkey, 21–23 July 2016; pp. 136–141. [Google Scholar]
- Saleem, G.; Akhtar, M.; Ahmed, N.; Qureshi, W.S. Automated analysis of visual leaf shape features for plant classification. Comput. Electron. Agric. 2019, 157, 270–280. [Google Scholar] [CrossRef]
- Turkoglu, M.; Hanbay, D. Leaf-based plant species recognition based on improved local binary pattern and extreme learning machine. Phys. Stat. Mech. Its Appl. 2019, 527, 121297. [Google Scholar] [CrossRef]
- Mostajer Kheirkhah, F.; Asghari, H. Plant leaf classification using GIST texture features. IET Comput. Vis. 2019, 13, 369–375. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, P. Plant species identification based on plant leaf using computer vision and machine learning techniques. J. Multimed. Inf. Syst. 2019, 6, 49–60. [Google Scholar] [CrossRef]
- Keivani, M.; Mazloum, J.; Sedaghatfar, E.; Tavakoli, M.B. Automated Analysis of Leaf Shape, Texture, and Color Features for Plant Classification. Trait. Signal 2020, 37, 17–28. [Google Scholar] [CrossRef]
- Rajesh, V.; Dudi, B.P. Performance analysis of leaf image classification using machine learning algorithms on different datasets. Turk. J. Physiother. Rehabil. 2021, 32, 2103–2108. [Google Scholar]
- Lee, S.H.; Chan, C.S.; Wilkin, P.; Remagnino, P. Deep-plant: Plant identification with convolutional neural networks. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 452–456. [Google Scholar]
- Liu, J.; Yang, S.; Cheng, Y.; Song, Z. Plant leaf classification based on deep learning. In Proceedings of the 2018 Chinese Automation Congress (CAC), Xi’an, China, 30 November–2 December 2018; pp. 3165–3169. [Google Scholar]
- Wei Tan, J.; Chang, S.W.; Abdul-Kareem, S.; Yap, H.J.; Yong, K.T. Deep learning for plant species classification using leaf vein morphometric. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 17, 82–90. [Google Scholar]
- Hu, J.; Chen, Z.; Yang, M.; Zhang, R.; Cui, Y. A multiscale fusion convolutional neural network for plant leaf recognition. IEEE Signal Process. Lett. 2018, 25, 853–857. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.S.; Catal, C.; Yalic, H.Y.; Temucin, H.; Tekinerdogan, B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Anubha Pearline, S.; Sathiesh Kumar, V.; Harini, S. A study on plant recognition using conventional image processing and deep learning approaches. J. Intell. Fuzzy Syst. 2019, 36, 1997–2004. [Google Scholar] [CrossRef]
- Riaz, S.A.; Naz, S.; Razzak, I. Multipath Deep Shallow Convolutional Networks for Large Scale Plant Species Identification in Wild Image. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–7. [Google Scholar]
- Litvak, M.; Divekar, S.; Rabaev, I. Urban Plants Classification Using Deep-Learning Methodology: A Case Study on a New Dataset. Signals 2022, 3, 524–534. [Google Scholar] [CrossRef]
- Arun, Y.; Viknesh, G. Leaf Classification for Plant Recognition Using EfficientNet Architecture. In Proceedings of the 2022 IEEE Fourth International Conference on Advances in Electronics, Computers and Communications (ICAECC), Bengaluru, India, 10–11 January 2022; pp. 1–5. [Google Scholar]
- Beikmohammadi, A.; Faez, K.; Motallebi, A. SWP-Leaf NET: A novel multistage approach for plant leaf identification based on deep learning. arXiv 2020, arXiv:2009.05139. [Google Scholar]
- Siddharth, S.C.; Singh, U.; Kaul, A.; Jain, S. A database of leaf images: Practice towards plant conservation with plant pathology. In Mendeley Data; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Wu, S.G.; Bao, F.S.; Xu, E.Y.; Wang, Y.X.; Chang, Y.F.; Xiang, Q.L. A leaf recognition algorithm for plant classification using probabilistic neural network. In Proceedings of the 2007 IEEE International Symposium on Signal Processing and Information Technology, Giza, Egypt, 15–18 December 2007; pp. 11–16. [Google Scholar]
- Munisami, T.; Ramsurn, M.; Kishnah, S.; Pudaruth, S. Plant leaf recognition using shape features and colour histogram with K-nearest neighbour classifiers. Procedia Comput. Sci. 2015, 58, 740–747. [Google Scholar] [CrossRef]
- Söderkvist, O.J.O. Computer Vision Classification of Leaves from Swedish Trees. Master’s Thesis, Linköping University, Linköping, Sweden, 2001. [Google Scholar]





| Dataset | Number of Classes | Total Number of Images | Training | Validation | Testing |
|---|---|---|---|---|---|
| Flavia Dataset | 32 | 1907 | 1323 (70%) | 178 (10%) | 406 (20%) |
| Folio Leaf Dataset | 32 | 637 | 445 (70%) | 62 (10%) | 130 (20%) |
| Swedish Leaf Dataset | 15 | 1125 | 675 (60%) | 105 (10%) | 345 (30%) |
| MalayaKew Leaf Dataset | 44 | 43,472 | 27,620 (80%) | 6952 (20%) | 8800 |
| Methods | Accuracy (%) |
|---|---|
| HOG with SVM [1] | 97.00 |
| C-SIFT with KD tree [1] | 98.00 |
| MSER with KD tree [1] | 90.00 |
| KNN [2] | 98.93 |
| ROM-LBP [3] | 98.94 |
| Cosine KNN [4] | 95.50 |
| SVM [4] | 89.90 |
| Patternnet Neural Network [4] | 72.20 |
| PBPSO with Decision Tree [6] | 98.58 |
| PBPSO with SVM [6] | 96.12 |
| PBPSO with Naive Bayes [6] | 92.01 |
| PBPSO with KNN [6] | 94.89 |
| Random Forest [7] | 84.11 |
| SVM [7] | 79.05 |
| Logistic Regression [7] | 84.11 |
| KNN [7] | 80.10 |
| Naive Bayes [7] | 72.25 |
| CNN [9] | 87.92 |
| D-Leaf with ANN [10] | 94.63 |
| VGG16 and LDA [12] | 99.10 |
| VGG16 [12] | 99.11 |
| CNN-RNN [12] | 99.11 |
| VGG19 with Logistic Regression [13] | 96.25 |
| SWP-LeafNet [17] | 99.67 |
| ViT | 99.75 |
| ResNet-50 | 98.28 |
| DenseNet-201 | 99.51 |
| Xception | 97.04 |
| Plant-CNN-ViT (proposed) | 100.00 |
| Methods | Accuracy (%) |
|---|---|
| PBPSO with Decision Tree [6] | 90.02 |
| PBPSO with SVM [6] | 88.02 |
| PBPSO with Naive Bayes [6] | 81.30 |
| PBPSO with KNN [6] | 89.21 |
| Random Forest [7] | 84.04 |
| SVM [7] | 60.63 |
| Logistic Regression [7] | 77.65 |
| KNN [7] | 70.21 |
| Naive Bayes [7] | 72.34 |
| VGG19 with Logistic Regression [13] | 96.53 |
| ViT | 96.92 |
| ResNet-50 | 96.15 |
| DenseNet-201 | 95.38 |
| Xception | 97.69 |
| Plant-CNN-ViT (proposed) | 100.00 |
| Methods | Accuracy (%) |
|---|---|
| OM-LBP [3] | 99.46 |
| GLCM with Multiclass-SVM [5] | 93.26 |
| Random Forest [7] | 84.61 |
| SVM [7] | 79.28 |
| Logistic Regression [7] | 84.02 |
| KNN [7] | 76.03 |
| Naive Bayes [7] | 73.07 |
| D-Leaf with ANN [10] | 98.09 |
| VGG19 with Logistic Regression [13] | 99.41 |
| ViT | 98.26 |
| ResNet-50 | 98.26 |
| DenseNet-201 | 98.55 |
| Xception | 95.07 |
| Plant-CNN-ViT (proposed) | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.P.; Lim, K.M.; Song, Y.X.; Alqahtani, A. Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer. Plants 2023, 12, 2642. https://doi.org/10.3390/plants12142642
Lee CP, Lim KM, Song YX, Alqahtani A. Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer. Plants. 2023; 12(14):2642. https://doi.org/10.3390/plants12142642
Chicago/Turabian StyleLee, Chin Poo, Kian Ming Lim, Yu Xuan Song, and Ali Alqahtani. 2023. "Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer" Plants 12, no. 14: 2642. https://doi.org/10.3390/plants12142642
APA StyleLee, C. P., Lim, K. M., Song, Y. X., & Alqahtani, A. (2023). Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer. Plants, 12(14), 2642. https://doi.org/10.3390/plants12142642





