qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer
Abstract
:1. Introduction
2. Results
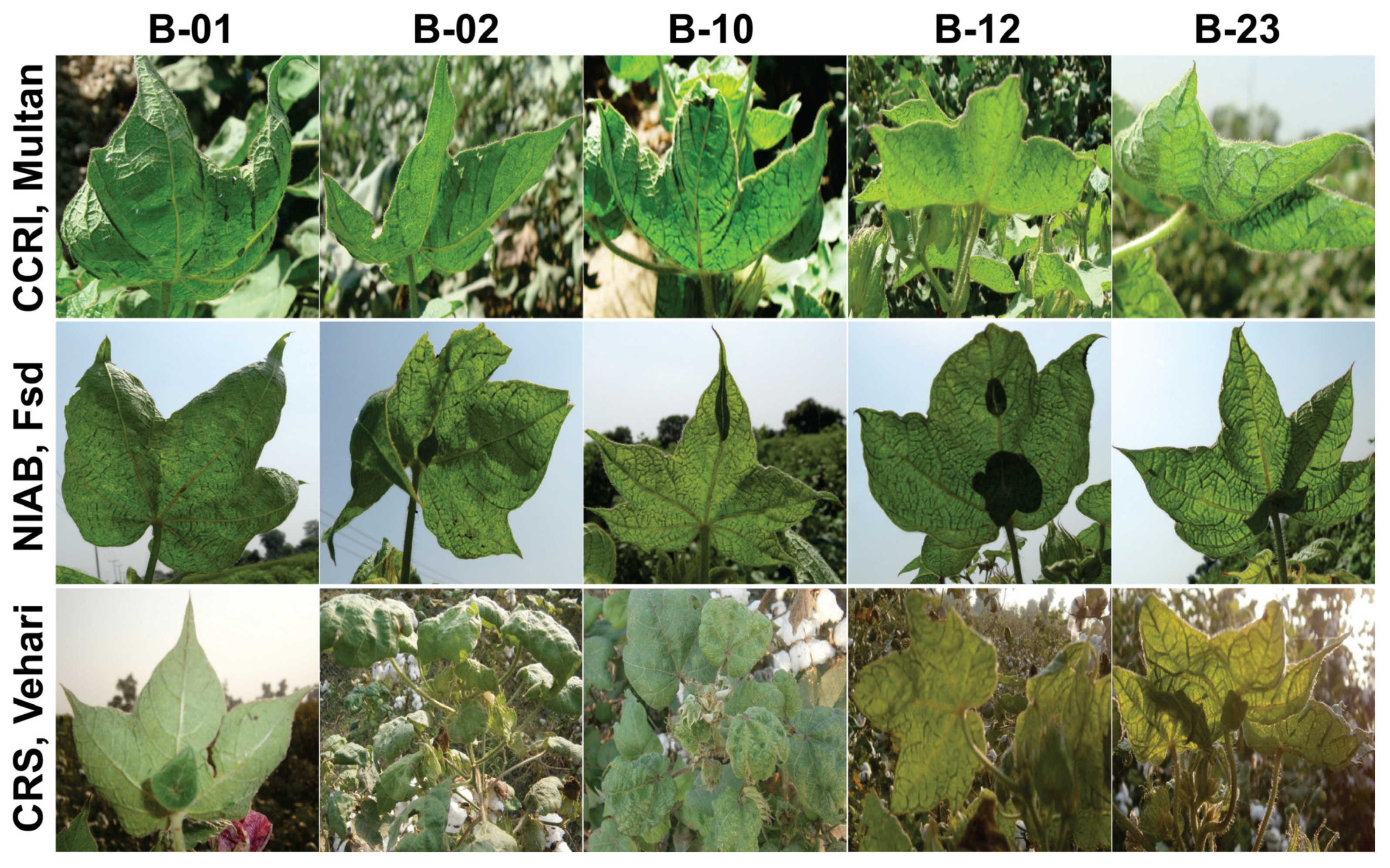
2.1. Phenotype and Disease Severity Index
2.2. Standard Curve and Quantification
2.3. Correlation between Virus Components and SCY
2.4. Path Coefficient Analysis
3. Discussion
4. Materials and Methods
4.1. Data Collection and DNA Extraction
4.2. Design of Primers
4.3. Preparation of Standard Curves for Quantification and Conditions for qPCR
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ur-Rahman, M.; Rahmat, Z.; Mahmood, A.; Abdullah, K.; Zafar, Y. Cotton Germplasm of Pakistan. In World Cotton Germplasm Resources; Ibrokhim, Y.A., Ed.; IntechOpen: Rijeka, Croatia, 2014; p. Ch. 6. [Google Scholar]
- FAOStat Food and Agriculture Organization of the United Nations Statistics Division. Available online: http://faostat3.fao.org/home/E (accessed on 25 April 2023).
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, M.Y.; Abdel-Salam, A.M.; Markham, P.G. Diversity of DNA 1; a satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, T.; Ali, M. A review of cotton diseases of Pakistan. Pak. Cotton 1975, 19, 71–86. [Google Scholar]
- Rishi, N.; Chauhan, M.S. Appearance of leaf curl disease of cotton in Northern India. J. Cotton Res. Dev. 1994, 8, 179–180. [Google Scholar]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liu, Y.; Robinson, D.J.; Harrison, B.D. Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 1998, 79, 915–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.; Hussain, D.; Malik, T.A.; Zafar, Y. Genetics of resistance against cotton leaf curl disease in Gossypium hirsutum. Plant Pathol. 2005, 54, 764–772. [Google Scholar] [CrossRef]
- Rahman, M.; Hussain, D.; Zafar, Y. Estimation of genetic divergence among elite cotton cultivars–genotypes by DNA fingerprinting technology. Crop Sci. 2002, 42, 2137–2144. [Google Scholar] [CrossRef]
- Amrao, L.; Amin, I.; Shahid, S.; Briddon, R.W.; Mansoor, S. Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Res. 2010, 152, 153–163. [Google Scholar] [CrossRef]
- Mahmood, T.; Arshad, M.; Gill, M.I.; Mahmood, H.T. Burewala strain of cotton leaf curl virus: A threat to CLCuV cotton resistant varieties. Asian J. Plant Sci. 2003, 2, 968–970. [Google Scholar] [CrossRef] [Green Version]
- Mansoor, S.; Briddon, R.W.; Zafar, Y.; Stanley, J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003, 8, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Zaidi, S.S.A.; Shakir, S.; Farooq, M.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Mansoor, S. Multiple begomoviruses found associated with cotton leaf curl disease in Pakistan in early 1990 are back in cultivated cotton. Sci. Rep. 2017, 7, 680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, S.; Budhauliya, R.; Das, B.; Gopalakrishnan, R.; Sharma, S.; Chatterjee, S.; Vanlalhmuaka; Raju, P.S.; Veer, V. Rebound of Cotton leaf curl Multan virus and its exclusive detection in cotton leaf curl disease outbreak, Punjab (India), 2015. Sci. Rep. 2017, 7, 17361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sattar, M.N.; Iqbal, Z.; Tahir, M.N.; Ullah, S. The prediction of a new CLCuD epidemic in the Old World. Front. Microbiol. 2017, 8, 631. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Amin, I.; Zaidi, S.S.; Rahman, S.U.; Farooq, M.; Fauquet, C.M.; Mansoor, S. Circular DNA enrichment sequencing reveals the viral/satellites genetic diversity associated with the third epidemic of cotton leaf curl disease. Biol. Methods Protoc. 2021, 6, bpab005. [Google Scholar] [CrossRef]
- Zubair, M.; Zaidi, S.; Shakir, S.; Amin, I.; Mansoor, S. An insight into Cotton leaf curl Multan betasatellite, the most important component of cotton leaf curl disease complex. Viruses 2017, 9, 280. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.; Mahmood, M.A.; Amin, I.; Mansoor, S. Geminiviruses also encode small proteins with specific functions. Trends Microbiol. 2021, 29, 1052–1054. [Google Scholar] [CrossRef]
- Varsani, A.; Martin, D.P.; Randles, J.W.; Vetten, H.J.; Thomas, J.E.; Fiallo-Olivé, E.; Navas-Castillo, J.; Lett, J.M.; Zerbini, F.M.; Roumagnac, P.; et al. Taxonomy update for the family Alphasatellitidae: New subfamily, genera, and species. Arch. Virol. 2021, 166, 3503–3511. [Google Scholar] [CrossRef]
- Wu, P.-J.; Zhou, X.-P. Interaction between a nanovirus-like component and the Tobacco curly shoot virus/satellite complex. Acta Biochim. Biophys. Sin. 2005, 37, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, Z.; Amin, I.; Mansoor, S.; Briddon, R.W. Effects of the transient expression of heterologous RNA virus-encoded silencing suppressors on the infectivity and systemic movement of tomato leaf curl New Delhi virus. Australas. Plant Pathol. 2020, 49, 531–540. [Google Scholar] [CrossRef]
- Abbas, Q.; Amin, I.; Mansoor, S.; Shafiq, M.; Wassenegger, M.; Briddon, R.W. The Rep proteins encoded by alphasatellites restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. VirusDisease 2017, 30, 101–105. [Google Scholar] [CrossRef]
- Zhao, L.; Che, X.; Wang, Z.; Zhou, X.; Xie, Y. Functional characterization of replication-associated proteins encoded by alphasatellites identified in Yunnan province, China. Viruses 2022, 14, 222. [Google Scholar] [CrossRef]
- Ashraf, K.H.; Hanif, M. Screening of cotton germplasm against cotton leaf curl virus. Pak. J. Bot. 2010, 42, 3327–3342. [Google Scholar]
- Khan, N.; Xing, F.; Feng, L.; Wang, Z.; Xin, M.; Xiong, S.; Wang, G.; Chen, H.; Du, W.; Li, Y. Comparative yield, fiber quality and dry matter production of cotton planted at various densities under equidistant row arrangement. Agronomy 2020, 10, 232. [Google Scholar] [CrossRef] [Green Version]
- Shafiq, M.; Iqbal, Z.; Ali, I.; Abbas, Q.; Mansoor, S.; Briddon, R.W.; Amin, I. Real-time quantitative PCR assay for the quantification of virus and satellites causing leaf curl disease in cotton in Pakistan. J. Virol. Methods 2017, 248, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Qazi, J.; Amin, I.; Mansoor, S.; Iqbal, J.; Briddon, R.W. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 2007, 128, 135–139. [Google Scholar] [CrossRef]
- Saeed, M.; Behjatnia, S.A.A.; Mansoor, S.; Zafar, Y.; Hasnain, S.; Rezaian, M.A. A single complementary-sense transcript of a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant-Microbe Interact. 2005, 18, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant-Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef] [Green Version]
- Amin, I.; Ilyas, M.; Mansoor, S.; Briddon, R.W.; Saeed, M. Role of DNA satellites in geminiviral disease complexes. In Emerging Geminiviral Diseases and Their Management; Sharma, P., Gaur, R.K., Ikegami, M., Eds.; Nova Science Publishers Inc.: New York, NY, USA, 2010; pp. 209–234. [Google Scholar]
- Iqbal, Z.; Sattar, M.N.; Kvarnheden, A.; Mansoor, S.; Briddon, R.W. Effects of the mutation of selected genes of Cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of Cotton leaf curl Multan betasatellite. Virus Res. 2012, 169, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M. Tomato leaf curl New Delhi virus DNA A component and Cotton leaf curl Multan betasatellite can cause mild transient symptoms in cotton. Acta Virol. 2010, 54, 317–318. [Google Scholar] [CrossRef]
- Eini, O.; Behjatnia, S.A.A.; Dogra, S.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Identification of sequence elements regulating promoter activity and replication of a monopartite begomovirus-associated DNA b satellite. J. Gen. Virol. 2009, 90, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.-H. bC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Wang, X.; Wu, J.; Briddon, R.W.; Zhou, X. βC1 encoded by tomato yellow leaf curl China betasatellite forms multimeric complexes in vitro and in vivo. Virology 2011, 409, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.P.; Usha, R.; Zrachya, A.; Levy, Y.; Spanov, H.; Gafni, Y. Protein-protein interactions and nuclear trafficking of coat protein and bC1 protein associated with Bhendi yellow vein mosaic disease. Virus Res. 2006, 122, 127–136. [Google Scholar] [CrossRef]
- Zhang, T.; Luan, J.-B.; Qi, J.-F.; Huang, C.-J.; Li, M.; Zhou, X.-P.; Liu, S.-S. Begomovirus–whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol. Ecol. 2012, 21, 1294–1304. [Google Scholar] [CrossRef]
- Iqbal, Z.; Shafiq, M.; Briddon, R. Cotton leaf curl Multan betasatellite impaired ToLCNDV ability to maintain cotton leaf curl Multan alphasatellite. Braz. J. Biol. 2022, 84, e260922. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Zheng, X.; Wei, X.; Nagalakshmi, U.; Hong, Y.; et al. Cotton leaf curl multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Markham, P.G. Clones of cotton leaf curl geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes 2000, 20, 19–26. [Google Scholar] [CrossRef]
- Fereres, A.; Moreno, A. Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Res. 2009, 141, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrode, S.D.; Bosque-Pérez, N.A.; Davis, T.S. Insect-borne plant pathogens and their vectors: Ecology, evolution, and complex interactions. Ann. Rev. Entomol. 2018, 63, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, M.; Sattar, M.N.; Shahid, M.S.; Al-Sadi, A.M.; Briddon, R.W. Interaction of watermelon chlorotic stunt virus with satellites. Australas. Plant Pathol. 2021, 50, 117–128. [Google Scholar] [CrossRef]
- Sattar, N.M.; Iqbal, Z.; Najabat Ali, S.; Amin, I.; Shafiq, M.; Khurshid, M. Natural occurrence of mesta yellow vein mosaic virus and DNA-satellites in ornamental sunflower (Helianthus spp.) in Pakistan. Saudi J. Biol. Sci. 2021, 28, 6621–6630. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A.; Wells, R. Dry matter allocation and fruiting patterns of cotton grown at two divergent plant populations. Crop Sci. 1997, 37, 797–802. [Google Scholar] [CrossRef]
- He, Z.; Zhang, D.; Mattison, C.P. Quantitative comparison of the storage protein distribution in glandless and glanded cottonseeds. Agric. Environ. Lett. 2022, 7, e20076. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Yang, L.; Chen, J.; Huang, C.; Liu, Y.; Jia, S.; Pan, L.; Zhang, D. Validation of a cotton-specific gene, Sad1, used as an endogenous reference gene in qualitative and real-time quantitative PCR detection of transgenic cottons. Plant Cell Rep. 2005, 24, 237–245. [Google Scholar] [CrossRef]
- Shafiq, M. Interactions of the Betasatellite Component of Begomovirus Complex; Pakistan Institute of Engineering and Applied Sciences: Islamabad, Pakistan, 2016. [Google Scholar]


| Variety | CCRI, Multan | NIAB, Faisalabad | CRS, Vehari | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Severity @ | Virus (ng·µg−1) | Beta. (ng·µg−1) | Alpha. (ng·µg−1) | SCY * (kg·ha−1) | Severity @ | Virus (ng·µg−1) | Beta. (ng·µg−1) | Alpha. (ng·µg−1) | SCY * (kg·ha−1) | Severity @ | Virus (ng·µg−1) | Beta. (ng·µg−1) | Alpha. (ng·µg−1) | SCY * (kg·ha−1) | |
| B-01 | 3 | 0.528 (±0.028) | 0.412 (±0.039) | 0.040 (±0.002) | 3118.0 (±100) | 3 | 0.008 (±0.0008) | 0.619 (±0.032) | 0.008 (±0.001) | 2302.0 (±100) | 2 | 0.009 (±0.0008) | 0.930 (±0.039) | 0.055 (±0.002) | 2165.0 (±100) |
| B-02 | 3 | 0.876 (±0.033) | 0.636 (±0.043) | 0.030 (±0.001) | 2875.0 (±100) | 2 | 0.006 (±0.0007) | 0.754 (±0.029) | 0.007 (±0.001) | 1704.0 (±100) | 3 | 0.103 (±0.0141) | 0.959 (±0.032) | 0.045 (±0.002) | 2082.0 (±100) |
| B-10 | 2 | 0.071 (±0.010) | 0.140 (±0.009) | 0.006 (±0.001) | 4054.0 (±100) | 3 | 0.063 (±0.0096) | 0.959 (±0.041) | 0.00001 (±0.0001) | 2474.0 (±100) | 2 | 0.972 (±0.0253) | 0.826 (±0.056) | 0.052 (±0.002) | 2209.0 (±100) |
| B-12 | 3 | 0.082 (±0.013) | 0.072 (±0.004) | 0.018 (±0.001) | 3159.0 (±100) | 2 | 0.020 (±0.0020) | 0.672 (±0.046) | 0.00001 (±0.0001) | 3805.0 (±100) | 3 | 0.139 (±0.0204) | 0.497 (±0.037) | 0.015 (±0.002) | 1282.0 (±100) |
| B-23 | 2 | 0.428 (±0.032) | 0.251 (±0.024) | 0.033 (±0.002) | 2960.0 (±100) | 3 | 0.313 (±0.0086) | 1.156 (±0.049) | 0.039 (±0.002) | 1936.0 (±100) | 1 | 0.060 (±0.0115) | 0.687 (±0.033) | 0.031 (±0.002) | 1296.0 (±100) |
| SCY | Begomovirus | Betasatellite | Alphasatellite | Symptom Severity | |
|---|---|---|---|---|---|
| SCY | 1 | ||||
| Begomovirus | 0.083 | 1 | |||
| Betasatellite | −0.540 * | 0.013 | 1 | ||
| Alphasatellite | −0.289 | 0.508 | 0.307 | 1 | |
| Symptom severity | 0.379 | −0.292 | −0.114 | −0.327 | 1 |
| Primer | Position | Sequence (5′-3′) | Amplicon Size (bp) |
|---|---|---|---|
| BegomoqPCRF1 | V2 | ATGTGGGATCCACTGTTAAATGAGTTCCC | 186 |
| BegomoqPCRR1 | GATTATATCTGCTGGTCGCTTCGACATAA | ||
| BetaqPCRF2 | βC1 | CAAGTATATCAAGTCTGTGAACTATATCTT | 194 |
| BetaqPCRR2 | GATACTATCCACAAAGTCACCATCGCTAAT | ||
| AlphaqPCRF2 | Rep | ATTCAAATTTCAAATTTGAAATCTTGGCA | 249 |
| AlphaqPCRR2 | CCTTCTTATCACGAGGGATATCAAATACAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, Z.; Shafiq, M.; Ali, S.; Mahmood, M.A.; Siddiqui, H.A.; Amin, I.; Briddon, R.W. qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer. Plants 2023, 12, 2645. https://doi.org/10.3390/plants12142645
Iqbal Z, Shafiq M, Ali S, Mahmood MA, Siddiqui HA, Amin I, Briddon RW. qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer. Plants. 2023; 12(14):2645. https://doi.org/10.3390/plants12142645
Chicago/Turabian StyleIqbal, Zafar, Muhammad Shafiq, Sajed Ali, Muhammad Arslan Mahmood, Hamid Anees Siddiqui, Imran Amin, and Rob W. Briddon. 2023. "qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer" Plants 12, no. 14: 2645. https://doi.org/10.3390/plants12142645






