Genome-Wide Characterization of PEBP Gene Family and Functional Analysis of TERMINAL FLOWER 1 Homologs in Macadamia integrifolia
Abstract
1. Introduction
2. Results
2.1. Identification of the PEBP Family in M. integrifolia
2.2. Gene Structure, Conserved Motif and Chromosomal Location Analysis of MiPEBP Genes
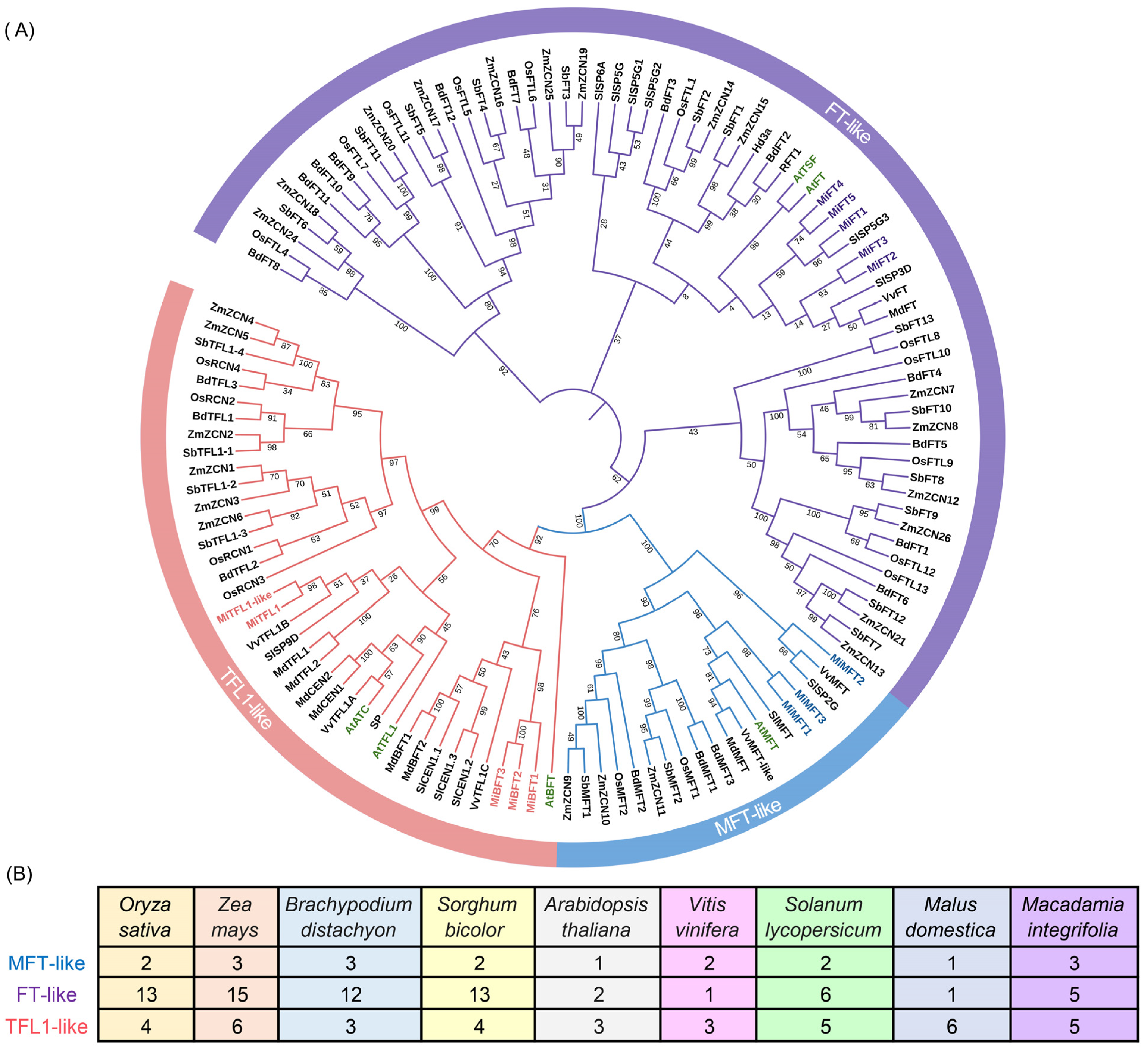
2.3. Phylogenetic Analysis and Classification of the PEBP Gene Family in M. integrifolia
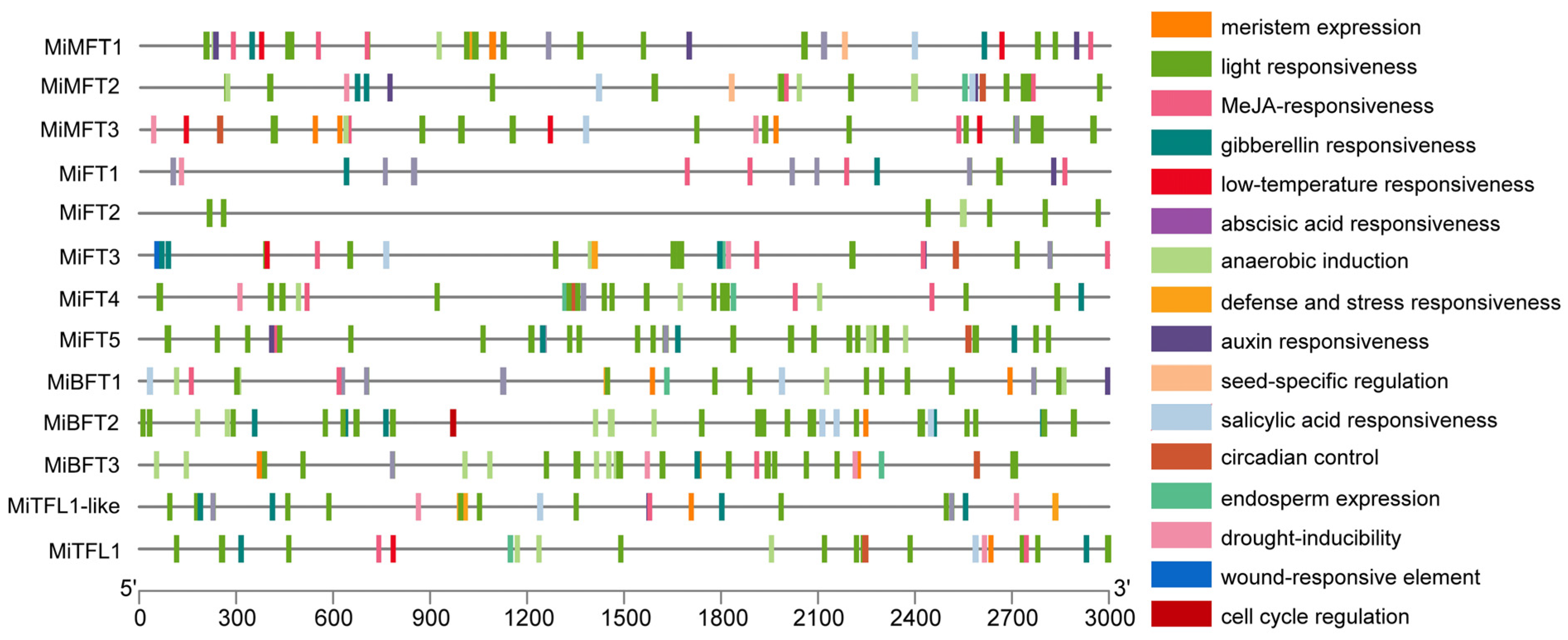
2.4. Analysis of Cis-Acting Regulatory Elements in Promoter Regions of MiPEBP Genes
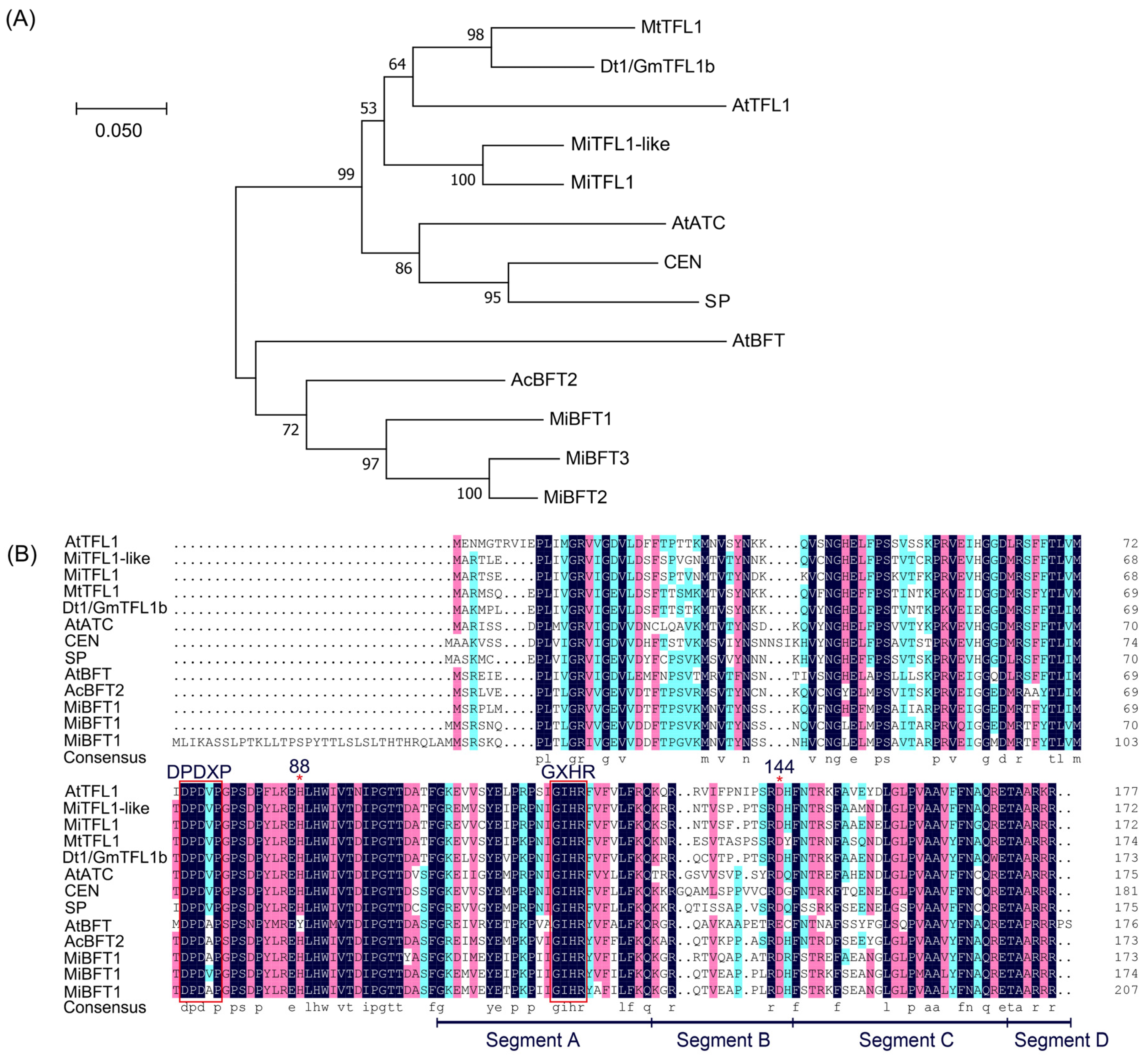
2.5. Identification and Multiple Alignment of MiTFL1 Homologue
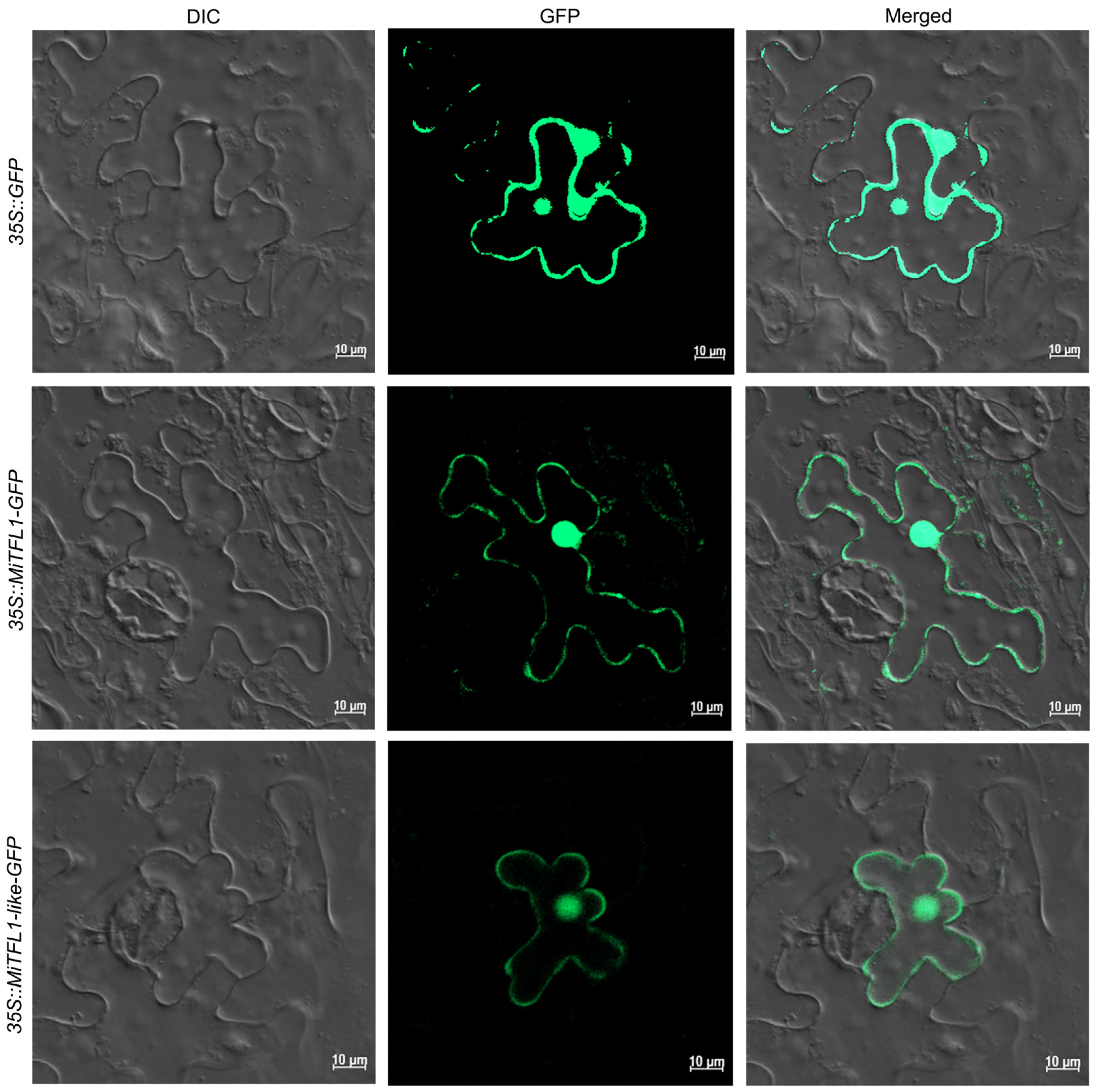
2.6. Subcellular Localizations of MiTFL1 and MiTFL1-like
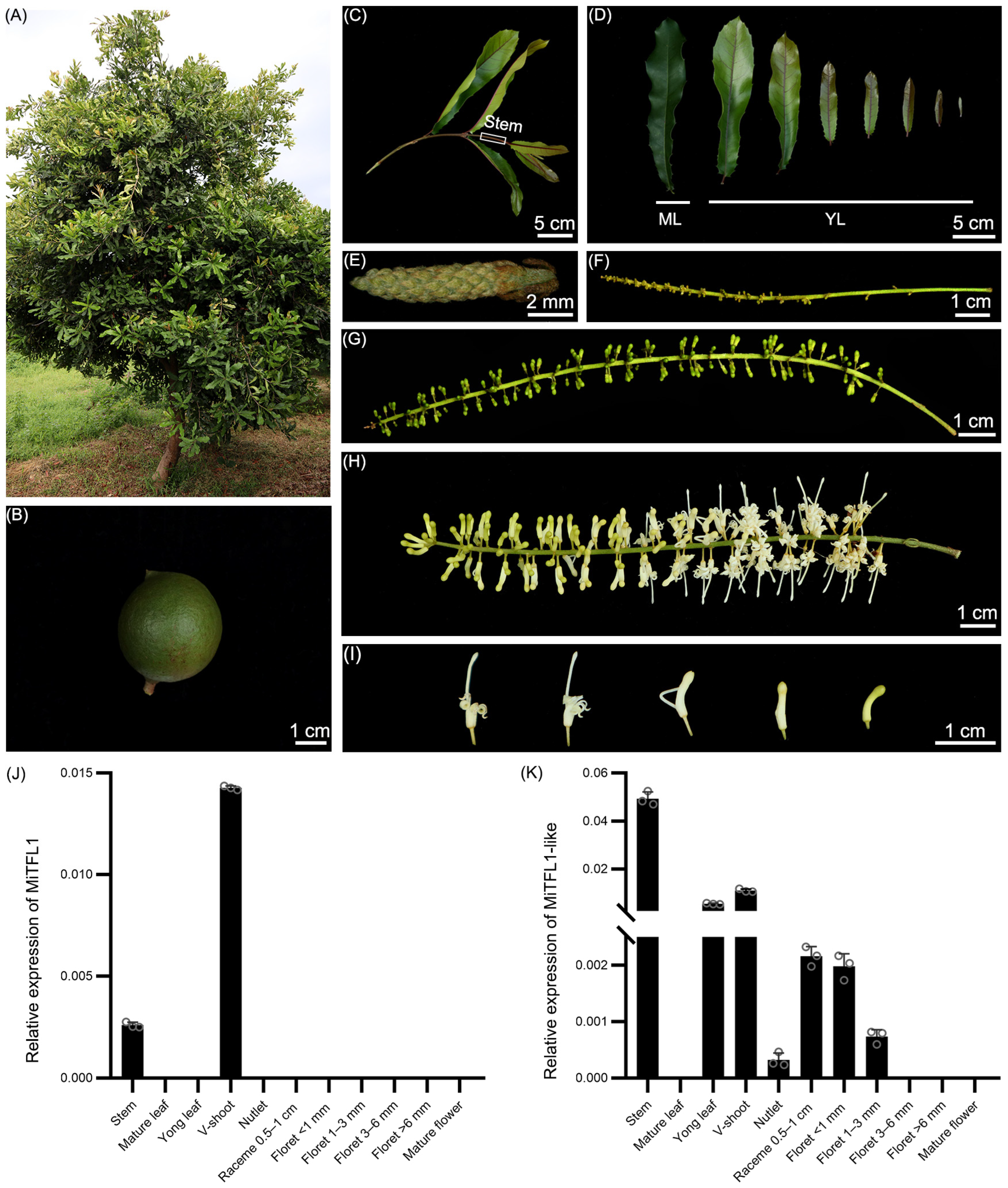
2.7. Expression Patterns of MiTFL1 and MiTFL1-like in M. integrifolia
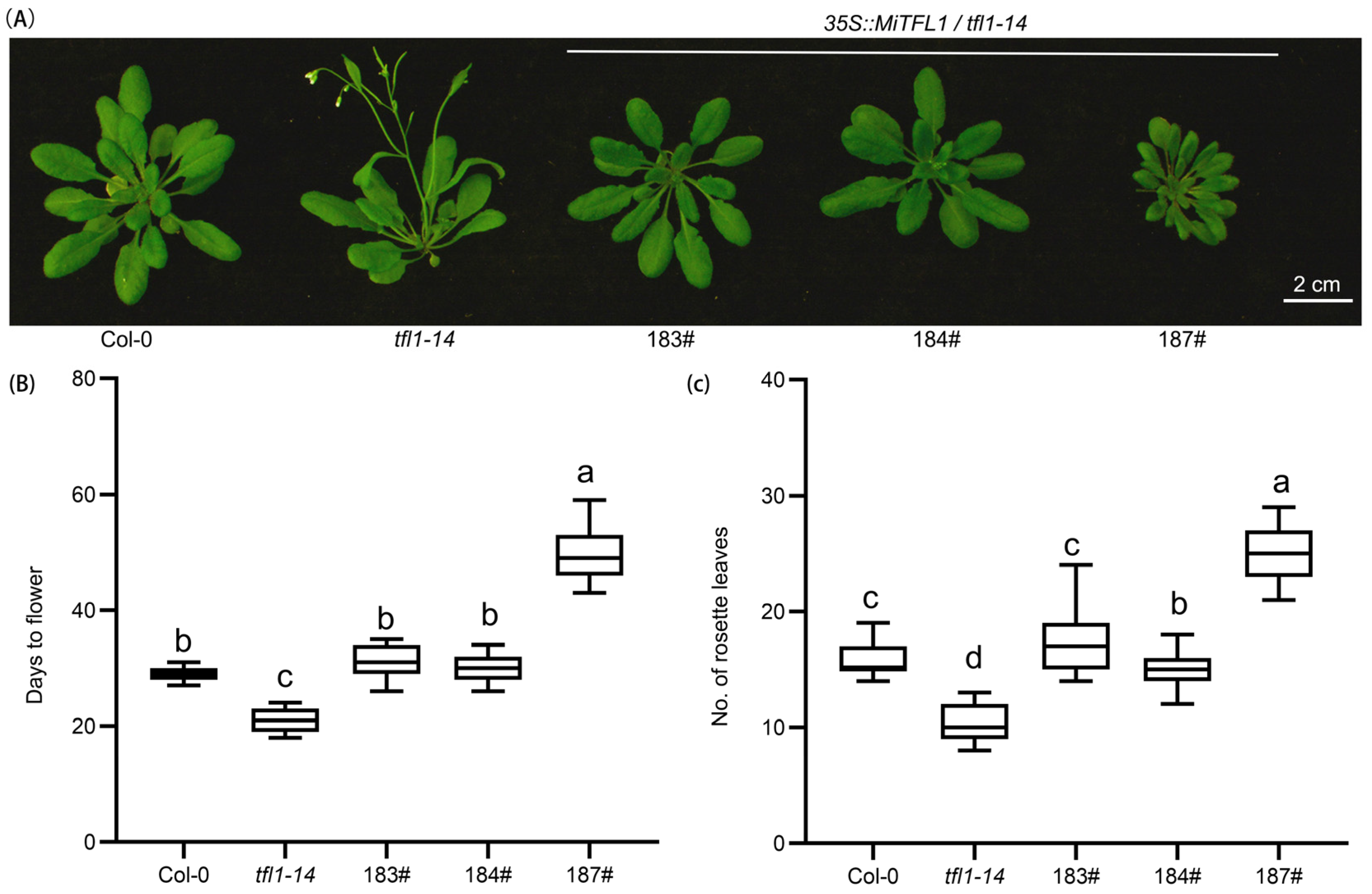
2.8. The Role of MiTFL1 in Regulating Flowering Time
2.9. The Phenotypic Analysis of MiTFL1 in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of PEBP Family Genes in M. integrifolia
4.3. Gene Structures, Protein Motifs and Chromosome Locations
4.4. Phylogenetic Analyses and Multiple Alignments
4.5. Analysis of Cis-Acting Regulatory Elements in Promoter of MiPEBPs
4.6. RNA Extraction and qRT-PCR
4.7. Vector Construction and Plant Transformation
4.8. Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoentgen, F.; Jolles, P. From structure to function: Possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett. 1995, 369, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Chautard, H.; Jacquet, M.; Schoentgen, F.; Bureaud, N.; Benedetti, H. Tfs1p, a member of the PEBP family, inhibits the Ira2p but not the Ira1p Ras GTPase-activating protein in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Karlgren, A.; Gyllenstrand, N.; Kallman, T.; Sundstrom, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef]
- Jin, S.; Nasim, Z.; Susila, H.; Ahn, J.H. Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Biol. 2021, 109, 20–30. [Google Scholar] [CrossRef]
- Hedman, H.; Kallman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Yang, K.Z.; Wei, X.X.; Wang, X.Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A Pair of Related Genes with Antagonistic Roles in Mediating Flowering Signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- So Yeon, Y.; Igor, K.; Jong Seob, L.; Detlef, W.; Ji Hoon, A. Acceleration of Flowering by Overexpression of MFT (MOTHER OF FT AND TFL1). Mol. Cells 2004, 17, 95–101. [Google Scholar]
- Vaistij, F.E.; Barros-Galvao, T.; Cole, A.F.; Gilday, A.D.; He, Z.; Li, Y.; Harvey, D.; Larson, T.R.; Graham, I.A. MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, 8442–8447. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal Behav. 2010, 5, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef]
- Huang, N.C.; Jane, W.N.; Chen, J.; Yu, T.S. Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J. Cell Mol. Biol. 2012, 72, 175–184. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Ando, E.; Ohnishi, M.; Wang, Y.; Matsushita, T.; Watanabe, A.; Hayashi, Y.; Fujii, M.; Ma, J.F.; Inoue, S.; Kinoshita, T. TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis. Plant Physiol. 2013, 162, 1529–1538. [Google Scholar] [CrossRef]
- Mimida, N.; Goto, K.; Kobayashi, Y.; Araki, T.; Ahn, J.H.; Weigel, D.; Murata, M.; Motoyoshi, F.; Sakamoto, W. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 2001, 6, 327–336. [Google Scholar] [CrossRef]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. Cell Mol. Biol. 2010, 63, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 2007, 19, 767–778. [Google Scholar] [CrossRef]
- Shannon, S.; Meeks-Wagner, D.R. A Mutation in the Arabidopsis TFL1 Gene Affects Inflorescence Meristem Development. Plant Cell 1991, 3, 877–892. [Google Scholar] [CrossRef]
- Ratcliffe, O.J.; Amaya, I.; Vincent, C.A.; Rothstein, S.; Carpenter, R.; Coen, E.S.; Bradley, D.J. A common mechanism controls the life cycle and architecture of plants. Development 1998, 125, 1609–1615. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef]
- Sun, L.; Nie, T.; Chen, Y.; Yin, Z. From Floral Induction to Blooming: The Molecular Mysteries of Flowering in Woody Plants. Int. J. Mol. Sci. 2022, 23, 10959. [Google Scholar] [CrossRef]
- Park, S.J.; Jiang, K.; Tal, L.; Yichie, Y.; Gar, O.; Zamir, D.; Eshed, Y.; Lippman, Z.B. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 2014, 46, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- Shalit, A.; Rozman, A.; Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y.; Lifschitz, E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 2009, 106, 8392–8397. [Google Scholar] [CrossRef] [PubMed]
- Gaston, A.; Potier, A.; Alonso, M.; Sabbadini, S.; Delmas, F.; Tenreira, T.; Cochetel, N.; Labadie, M.; Prevost, P.; Folta, K.M.; et al. The FveFT2 florigen/FveTFL1 antiflorigen balance is critical for the control of seasonal flowering in strawberry while FveFT3 modulates axillary meristem fate and yield. New Phytol. 2021, 232, 372–387. [Google Scholar] [CrossRef]
- Perilleux, C.; Bouche, F.; Randoux, M.; Orman-Ligeza, B. Turning Meristems into Fortresses. Trends Plant Sci. 2019, 24, 431–442. [Google Scholar] [CrossRef]
- Li, C.; Fu, Q.; Niu, L.; Luo, L.; Chen, J.; Xu, Z.F. Three TFL1 homologues regulate floral initiation in the biofuel plant Jatropha curcas. Sci. Rep. 2017, 7, 43090. [Google Scholar] [CrossRef]
- Freiman, A.; Shlizerman, L.; Golobovitch, S.; Yablovitz, Z.; Korchinsky, R.; Cohen, Y.; Samach, A.; Chevreau, E.; Le Roux, P.M.; Patocchi, A.; et al. Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2. Planta 2012, 235, 1239–1251. [Google Scholar] [CrossRef]
- Kotoda, N.; Iwanami, H.; Takahashi, S.; Abe, K. Antisense Expression of MdTFL1, a TFL1-like Gene, Reduces the Juvenile Phase in Apple. J. Am. Soc. Hortic. Sci. JASHS 2006, 131, 74–81. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Trankner, C.; Hanke, M.V. The MdTFL1 gene of apple (Malus x domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. Cell Mol. Biol. 2012, 69, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.; Wang, C.T.; Ma, C.; Shevchenko, O.; Dye, S.J.; Puzey, J.R.; Etherington, E.; Sheng, X.; Meilan, R.; Strauss, S.H.; et al. Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. Cell Mol. Biol. 2010, 62, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Li, G.; Ni, S.; He, X.; Zheng, C.; Liu, Z.; Gong, L.; Kong, G.; Li, W.; Liu, J. The Chromosome-Scale Reference Genome of Macadamia tetraphylla Provides Insights into Fatty Acid Biosynthesis. Front. Genet. 2022, 13, 835363. [Google Scholar] [CrossRef]
- Topp, B.L.; Nock, C.J.; Hardner, C.M.; Alam, M.; O’Connor, K.M. Macadamia (Macadamia spp.) Breeding. In Advances in Plant Breeding Strategies: Nut and Beverage Crops; Springer: Berlin/Heidelberg, Germany, 2019; pp. 221–251. [Google Scholar]
- Hu, W.; Fitzgerald, M.; Topp, B.; Alam, M.; O’Hare, T.J. A review of biological functions, health benefits, and possible de novo biosynthetic pathway of palmitoleic acid in macadamia nuts. J. Funct. Foods 2019, 62, 103520. [Google Scholar] [CrossRef]
- Shuai, X.; Dai, T.; Chen, M.; Liang, R.; Du, L.; Chen, J.; Liu, C. Comparative Study of Chemical Compositions and Antioxidant Capacities of Oils Obtained from 15 Macadamia (Macadamia integrifolia) Cultivars in China. Foods 2021, 10, 1031. [Google Scholar] [CrossRef]
- Chagné, D. Chapter One—Whole Genome Sequencing of Fruit Tree Species. In Advances in Botanical Research; Plomion, C., Adam-Blondon, A.-F., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 74, pp. 1–37. [Google Scholar]
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; Furtado, A.; Henry, R.J.; King, G.J. Chromosome-Scale Assembly and Annotation of the Macadamia Genome (Macadamia integrifolia HAES 741). G3 Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, W.; Zhang, X.; Ma, X.; Zhang, S.; Chen, S.; Wang, Y.; Jia, H.; Liao, Z.; Lin, J.; et al. Signatures of selection in recently domesticated macadamia. Nat. Commun. 2022, 13, 242. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, X.; Nie, M.; Li, L.; Luo, Y.; Fan, Y.; Liu, C.; Ye, X.; Zou, L. Genome-wide identification and expression analysis disclose the pivotal PHOSPHATIDYLETHANOLAMINE BINDING PROTEIN members that may be utilized for yield improvement of Chenopodium quinoa. Front. Plant Sci. 2022, 13, 1119049. [Google Scholar] [CrossRef]
- Xu, H.; Guo, X.; Hao, Y.; Lu, G.; Li, D.; Lu, J.; Zhang, T. Genome-wide characterization of PEBP gene family in Perilla frutescens and PfFT1 promotes flowering time in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1026696. [Google Scholar] [CrossRef]
- Zhao, C.; Zhu, M.; Guo, Y.; Sun, J.; Ma, W.; Wang, X. Genomic Survey of PEBP Gene Family in Rice: Identification, Phylogenetic Analysis, and Expression Profiles in Organs and under Abiotic Stresses. Plants 2022, 11, 1576. [Google Scholar] [CrossRef] [PubMed]
- Shemesh-Mayer, E.; Faigenboim, A.; Ben Michael, T.E.; Kamenetsky-Goldstein, R. Integrated Genomic and Transcriptomic Elucidation of Flowering in Garlic. Int. J. Mol. Sci. 2022, 23, 13876. [Google Scholar] [CrossRef] [PubMed]
- Venail, J.; da Silva Santos, P.H.; Manechini, J.R.; Alves, L.C.; Scarpari, M.; Falcao, T.; Romanel, E.; Brito, M.; Vicentini, R.; Pinto, L.; et al. Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS T orthologue in sugarcane. J. Exp. Bot. 2022, 73, 2035–2049. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16 (Suppl. S1), S18–S31. [Google Scholar] [CrossRef] [PubMed]
- Freytes, S.N.; Canelo, M.; Cerdan, P.D. Regulation of Flowering Time: When and Where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef]
- Luccioni, L.; Krzymuski, M.; Sanchez-Lamas, M.; Karayekov, E.; Cerdan, P.D.; Casal, J.J. CONSTANS delays Arabidopsis flowering under short days. Plant J. Cell Mol. Biol. 2019, 97, 923–932. [Google Scholar] [CrossRef]
- Collani, S.; Neumann, M.; Yant, L.; Schmid, M. FT Modulates Genome-Wide DNA-Binding of the bZIP Transcription Factor FD. Plant Physiol. 2019, 180, 367–380. [Google Scholar] [CrossRef]
- Bradley, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Rothstein, S.; Coen, E. Control of inflorescence architecture in Antirrhinum. Nature 1996, 379, 791–797. [Google Scholar] [CrossRef]
- Nakagawa, M.; Shimamoto, K.; Kyozuka, J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002, 29, 743–750. [Google Scholar] [CrossRef]
- Liu, C.; Teo, Z.W.; Bi, Y.; Song, S.; Xi, W.; Yang, X.; Yin, Z.; Yu, H. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 2013, 24, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Watanabe, S.; Uchiyama, T.; Kong, F.; Kanazawa, A.; Xia, Z.; Nagamatsu, A.; Arai, M.; Yamada, T.; Kitamura, K.; et al. The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010, 153, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Klasfeld, S.; Wagner, D. Molecular regulation of plant developmental transitions and plant architecture via PEPB family proteins: An update on mechanism of action. J. Exp. Bot. 2021, 72, 2301–2311. [Google Scholar] [CrossRef]
- Wen, C.; Zhao, W.; Liu, W.; Yang, L.; Wang, Y.; Liu, X.; Xu, Y.; Ren, H.; Guo, Y.; Li, C.; et al. CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 2019, 146, dev180166. [Google Scholar] [CrossRef]
- Alam, M.M.; Howell, E.; Hardner, C.M.; Topp, B.L. Variation in precocity in a macadamia breeding population. In International Symposia on Tropical and Temperate Horticulture-Istth2016; ISHS: Cairns, Queensland, Australia, 2018; Volume 1205, pp. 645–651. [Google Scholar] [CrossRef]
- O’Connor, K.M.; Hardner, C.M.; Alam, M.M.; Hayes, B.J.; Topp, B.L. Variation in floral and growth traits in a macadamia breeding population. In International Symposia on Tropical and Temperate Horticulture-Istth2016; ISHS: Cairns, Queensland, Australia, 2018; Volume 1205, pp. 623–630. [Google Scholar] [CrossRef]
- Toft, B.D.; Alam, M.M.; Topp, B.L. Broad-sense heritability and inter-trait relationships in young macadamia architecture, flowering and yield. In International Symposia on Tropical and Temperate Horticulture-Istth2016; ISHS: Cairns, Queensland, Australia, 2018; Volume 1205, pp. 609–616. [Google Scholar] [CrossRef]
- O’Connor, K.; Hayes, B.; Hardner, C.; Nock, C.; Baten, A.; Alam, M.; Henry, R.; Topp, B. Genome-wide association studies for yield component traits in a macadamia breeding population. BMC Genom. 2020, 21, 199. [Google Scholar] [CrossRef]
- O’Connor, K.; Hayes, B.; Topp, B. Prospects for increasing yield in macadamia using component traits and genomics. Tree Genet. Genomes 2018, 14, 7. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]


| Gene Subfamily | Gene ID | Transcript ID | Sequence ID | Number of Amino Acid | Molecular Weight (MW) | Theoretical Isoelectric Point (PI) | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|
| MFT-like | MiMFT1 | MiMFT1 | XP_042502857.1 | 172 | 18.98 | 9.16 | −0.058 |
| MiMFT2 | MiMFT2 | XP_042520767.1 | 180 | 19.69 | 9.42 | −0.236 | |
| MiMFT3 | MiMFT3.1 | XP_042509881.1 | 203 | 22.66 | 5.87 | −0.158 | |
| MiMFT3.2 | XP_042509888.1 | 172 | 19.03 | 7.02 | −0.132 | ||
| FT-like | MiFT1 | MiFT1.1 | XP_042482853.1 | 190 | 21.73 | 9.4 | −0.464 |
| MiFT1.2 | XP_042482854.1 | 183 | 20.83 | 9 | −0.483 | ||
| MiFT2 | MiFT2 | XP_042494244.1 | 174 | 19.71 | 6.12 | −0.402 | |
| MiFT3 | MiFT3.1 | XP_042503696.1 | 174 | 19.74 | 7.95 | −0.304 | |
| MiFT3.2 | XP_042503697.1 | 174 | 19.8 | 9.41 | −0.347 | ||
| MiFT4 | MiFT4 | XP_042516980.1 | 185 | 20.96 | 7.68 | −0.263 | |
| MiFT5 | MiFT5 | XP_042517052.1 | 183 | 20.983 | 8.49 | −0.425 | |
| TFL1-like | MiBFT1 | MiBFT1 | XP_042506411.1 | 173 | 19.58 | 9.47 | −0.279 |
| MiBFT2 | MiBFT2 | XP_042510995.1 | 174 | 19.62 | 7.89 | −0.352 | |
| MiBFT3 | MiBFT3 | XP_042511022.1 | 207 | 23.12 | 9.41 | −0.286 | |
| MiTFL1-like | MiTFL1-like | XP_042489543.1 | 172 | 19.42 | 9.18 | −0.273 | |
| MiTFL1 | MiTFL1 | XP_042494365.1 | 172 | 19.44 | 8.66 | −0.281 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Ning, C.; Liu, Z.; Zheng, C.; Mao, Y.; Wu, Q.; Wang, D.; Liu, M.; Zhou, S.; Yang, L.; et al. Genome-Wide Characterization of PEBP Gene Family and Functional Analysis of TERMINAL FLOWER 1 Homologs in Macadamia integrifolia. Plants 2023, 12, 2692. https://doi.org/10.3390/plants12142692
Yang J, Ning C, Liu Z, Zheng C, Mao Y, Wu Q, Wang D, Liu M, Zhou S, Yang L, et al. Genome-Wide Characterization of PEBP Gene Family and Functional Analysis of TERMINAL FLOWER 1 Homologs in Macadamia integrifolia. Plants. 2023; 12(14):2692. https://doi.org/10.3390/plants12142692
Chicago/Turabian StyleYang, Jing, Conghui Ning, Ziyan Liu, Cheng Zheng, Yawen Mao, Qing Wu, Dongfa Wang, Mingli Liu, Shaoli Zhou, Liling Yang, and et al. 2023. "Genome-Wide Characterization of PEBP Gene Family and Functional Analysis of TERMINAL FLOWER 1 Homologs in Macadamia integrifolia" Plants 12, no. 14: 2692. https://doi.org/10.3390/plants12142692
APA StyleYang, J., Ning, C., Liu, Z., Zheng, C., Mao, Y., Wu, Q., Wang, D., Liu, M., Zhou, S., Yang, L., He, L., Liu, Y., He, C., Chen, J., & Liu, J. (2023). Genome-Wide Characterization of PEBP Gene Family and Functional Analysis of TERMINAL FLOWER 1 Homologs in Macadamia integrifolia. Plants, 12(14), 2692. https://doi.org/10.3390/plants12142692






