Long-Distance Movement of Solanum tuberosum Translationally Controlled Tumor Protein (StTCTP) mRNA
Abstract
:1. Introduction
2. Results
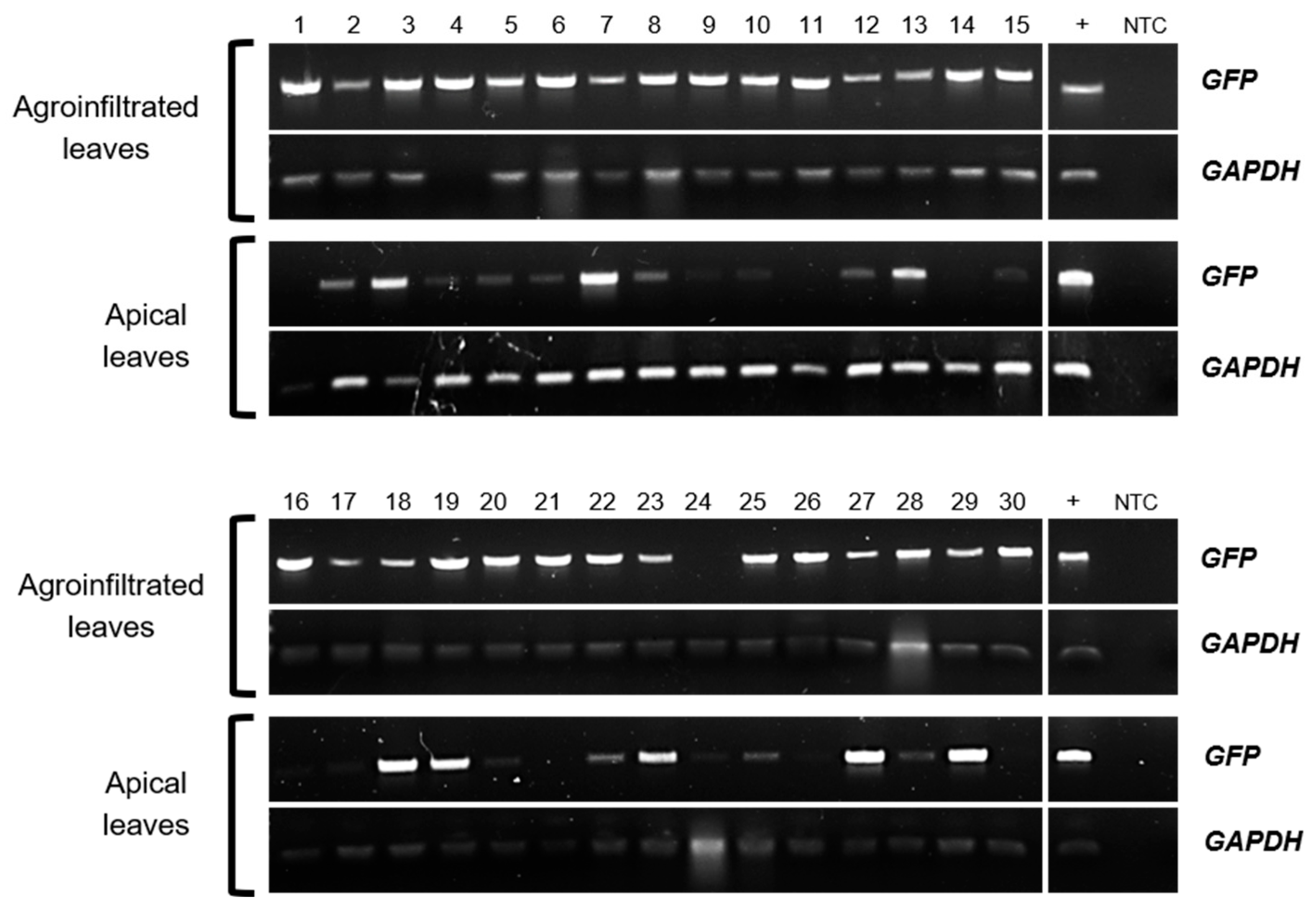
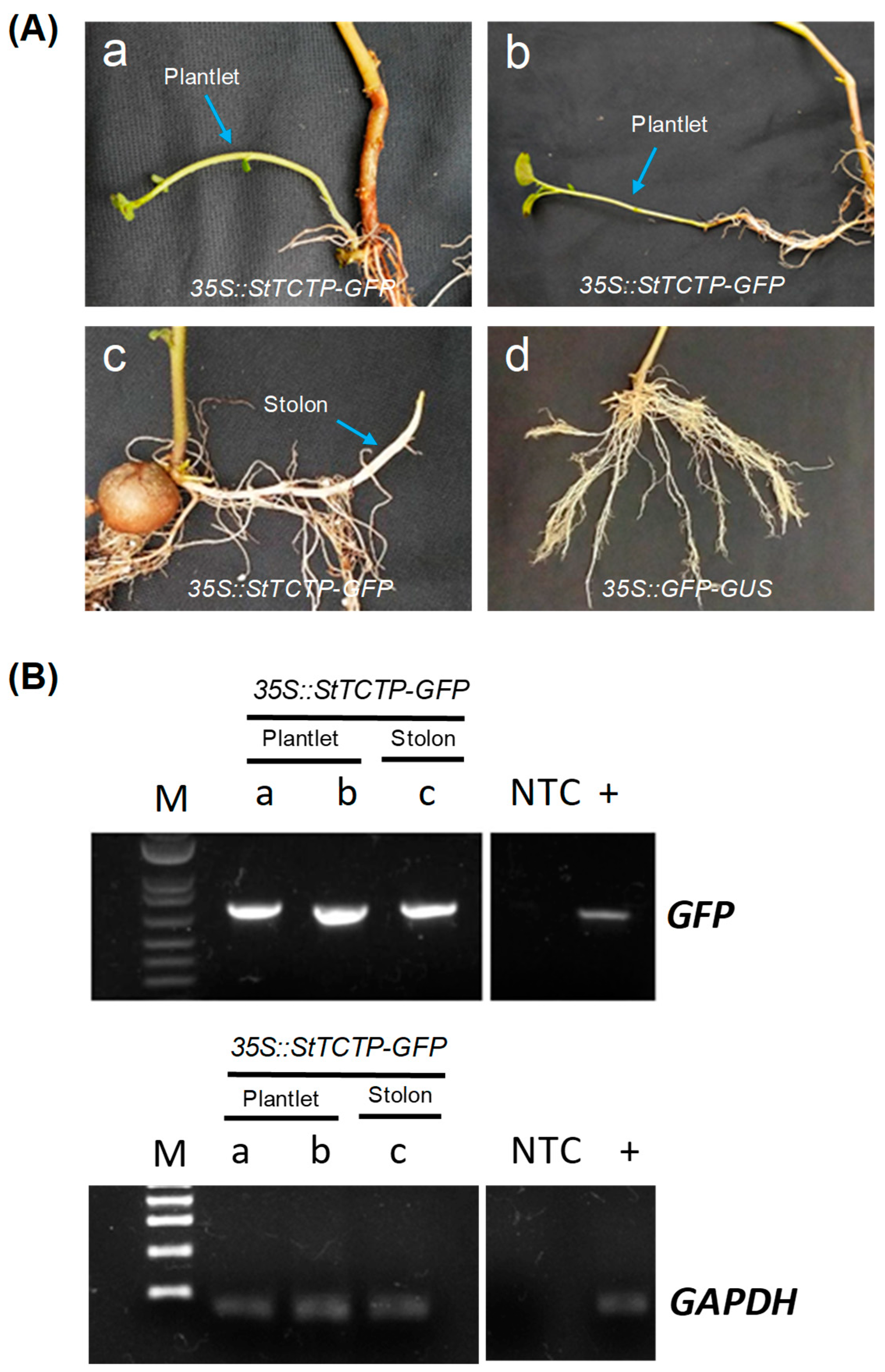
2.1. GFP-Fused StTCTP Transcript Is Transported Long Distance
2.2. Phenotype of Plants Agroinfiltrated with 35S::StTCTP-GFP or 35S::GFP-GUS
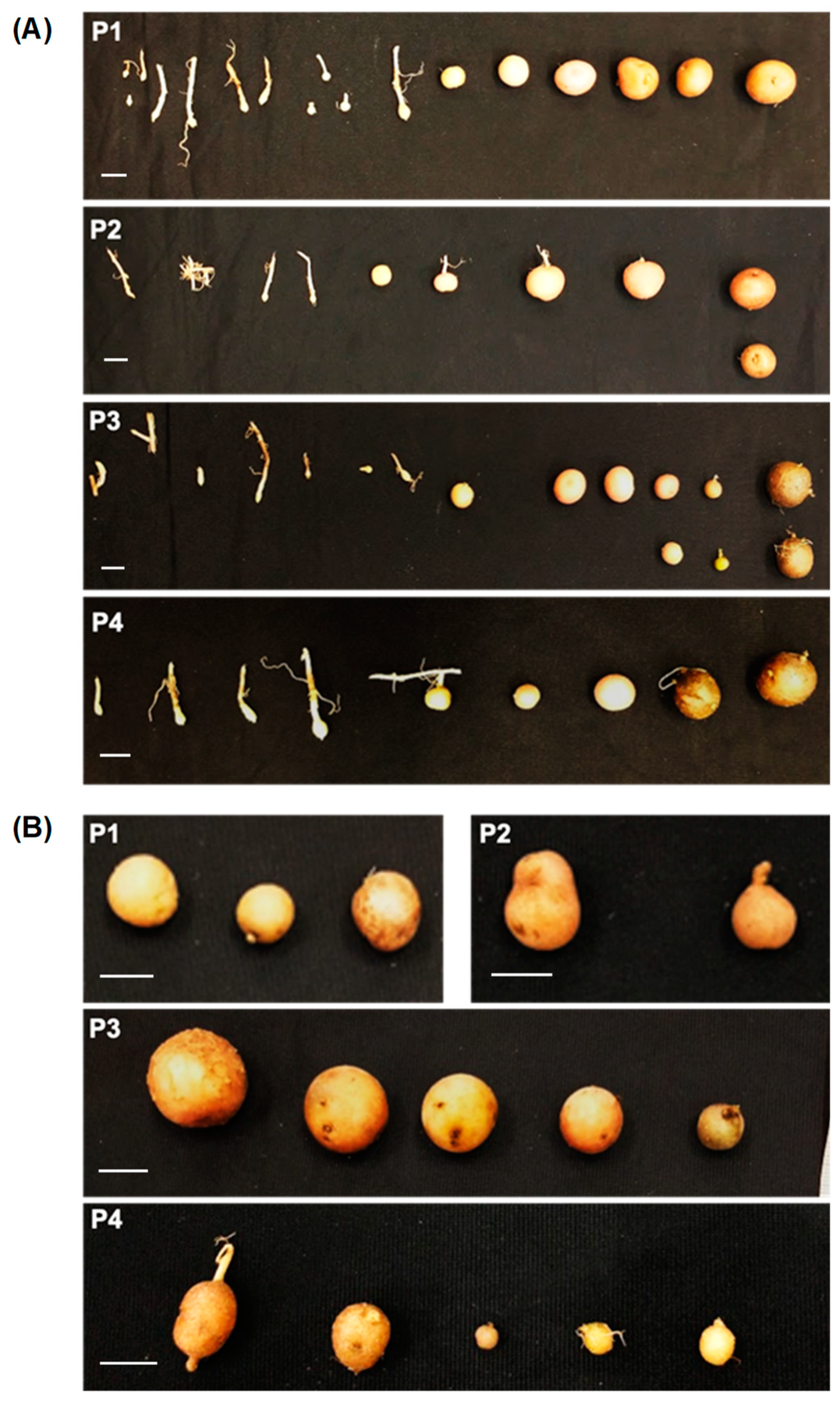
2.2.1. StTCTP Induced an Increase in the Number of Tubers in Early Stages of Tuberization
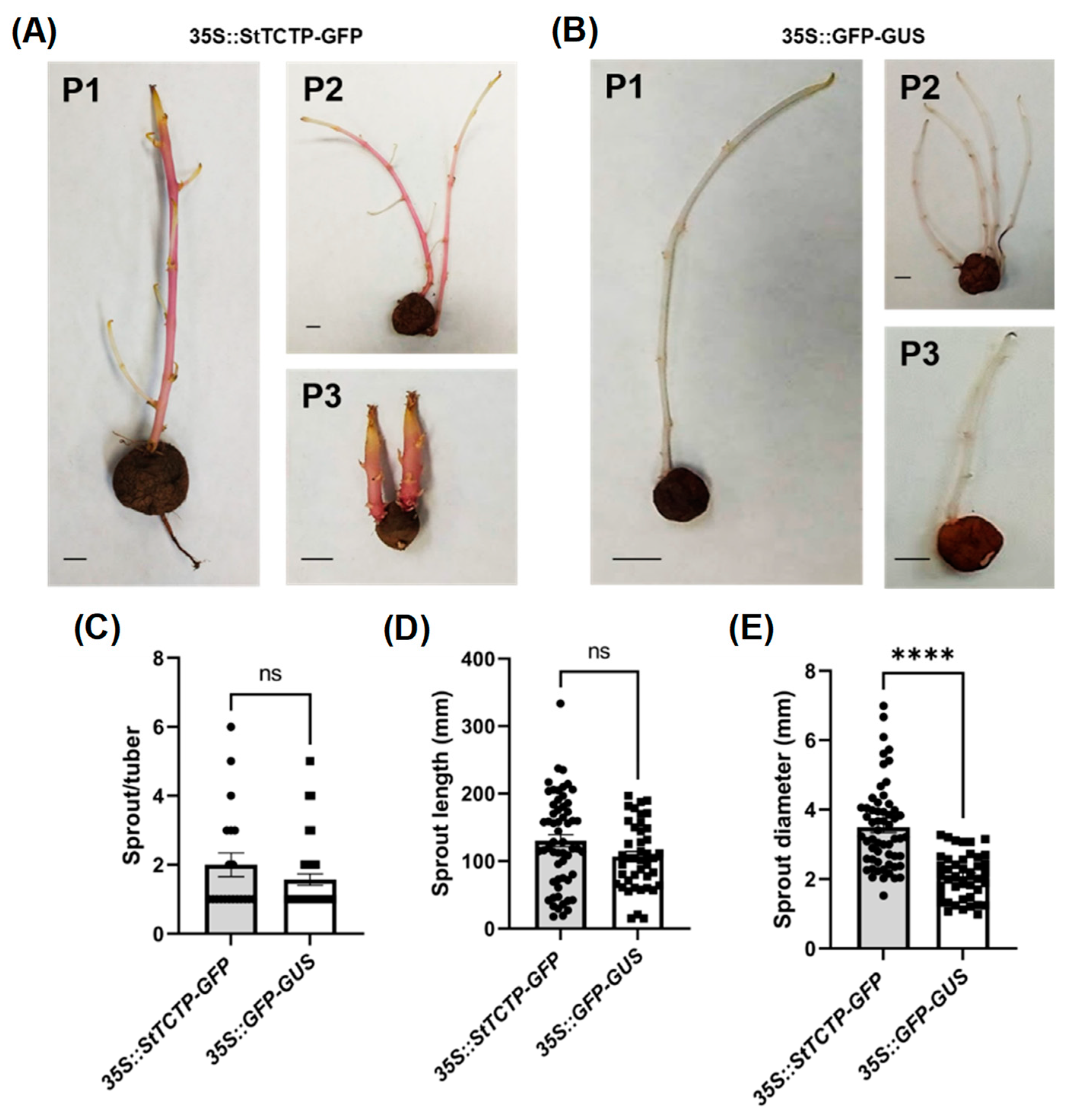
2.2.2. StTCTP Promoted an Increase in Sprout Diameter
2.3. Molecular Docking Simulation Indicated Interaction between StTCTP and PTB1/6
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction and Vector Construction
4.3. Potato Transient Transformation
4.4. Endpoint RT-PCR
4.5. Quantitative RT-PCR (RT-qPCR)
4.6. Shoot Induction Assay
4.7. Prediction of Tertiary Structures
4.7.1. Prediction of 3D Structures of mRNA
4.7.2. Prediction of 3D Structures of Proteins
4.7.3. Protein-RNA Interactions by Molecular Docking Simulation
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ruiz-Medrano, R.; Xoconostle-Cázares, B.; Lucas, W.J. Phloem Long-Distance Transport of CmNACP MRNA: Implications for Supracellular Regulation in Plants. Development 1999, 126, 4405–4419. [Google Scholar] [CrossRef] [PubMed]
- Omid, A.; Keilin, T.; Glass, A.; Leshkowitz, D.; Wolf, S. Characterization of Phloem-Sap Transcription Profile in Melon Plants. J. Exp. Bot. 2007, 58, 3645–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Medrano, R.; Moya, J.H.; Xoconostle-Cázares, B.; Lucas, W.J. Influence of Cucumber Mosaic Virus Infection on the MRNA Population Present in the Phloem Translocation Stream of Pumpkin Plants. Funct. Plant Biol. 2007, 34, 292. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; LeBlanc, M.L.; Wafula, E.K.; dePamphilis, C.W.; Westwood, J.H. Genomic-Scale Exchange of mRNA between a Parasitic Plant and Its Hosts. Science 2014, 345, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Kim, G. RNA Mobility in Parasitic Plant–Host Interactions. RNA Biol. 2017, 14, 450–455. [Google Scholar] [CrossRef] [Green Version]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis Messenger RNAs Transported to Distant Tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef]
- Chen, Y.; Singh, A.; Kaithakottil, G.G.; Mathers, T.C.; Gravino, M.; Mugford, S.T.; Van Oosterhout, C.; Swarbreck, D.; Hogenhout, S.A. An Aphid RNA Transcript Migrates Systemically within Plants and Is a Virulence Factor. Proc. Natl. Acad. Sci. USA 2020, 117, 12763–12771. [Google Scholar] [CrossRef] [PubMed]
- Doering-Saad, C.; Newbury, H.; Couldridge, C.; Bale, J.; Pritchard, J. A Phloem-Enriched CDNA Library from Ricinus: Insights into Phloem Function. J. Exp. Bot. 2006, 57, 3183–3193. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Medina, C.; Atkins, C.A.; Mann, A.J.; Jordan, M.E.; Smith, P.M. Macromolecular Composition of Phloem Exudate from White Lupin (Lupinus albus L.). BMC Plant Biol. 2011, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Mao, L.; Jittayasothorn, Y.; Kang, Y.; Jiao, C.; Fei, Z.; Zhong, G.-Y. Messenger RNA Exchange between Scions and Rootstocks in Grafted Grapevines. BMC Plant Biol. 2015, 15, 251. [Google Scholar] [CrossRef] [Green Version]
- Haywood, V.; Yu, T.-S.; Huang, N.-C.; Lucas, W.J. Phloem Long-Distance Trafficking of GIBBERELLIC ACID-INSENSITIVE RNA Regulates Leaf Development: Phloem Delivery of RNA Regulates Leaf Development. Plant J. 2005, 42, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, B.; Kondhare, K.R.; Hannapel, D.J.; Banerjee, A.K. Mobile RNAs and Proteins: Prospects in Storage Organ Development of Tuber and Root Crops. Plant Sci. 2019, 284, 73–81. [Google Scholar] [CrossRef]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem-Mobile Aux/IAA Transcripts Target to the Root Tip and Modify Root Architecture. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.-R. MicroRNA399 Is a Long-Distance Signal for the Regulation of Plant Phosphate Homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zheng, Y.; Ham, B.-K.; Chen, J.; Yoshida, A.; Kochian, L.V.; Fei, Z.; Lucas, W.J. Vascular-Mediated Signalling Involved in Early Phosphate Stress Response in Plants. Nat. Plants 2016, 2, 16033. [Google Scholar] [CrossRef]
- Yoo, B.-C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A Systemic Small RNA Signaling System in Plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef] [Green Version]
- Champigny, M.J.; Isaacs, M.; Carella, P.; Faubert, J.; Fobert, P.R.; Cameron, R.K. Long Distance Movement of DIR1 and Investigation of the Role of DIR1-like during Systemic Acquired Resistance in Arabidopsis. Front. Plant Sci. 2013, 4, 230. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Adam, H.; Díaz-Mendoza, M.; Żurczak, M.; González-Schain, N.D.; Suárez-López, P. Graft-Transmissible Induction of Potato Tuberization by the MicroRNA MiR172. Development 2009, 136, 2873–2881. [Google Scholar] [CrossRef] [Green Version]
- Navarro, C.; Abelenda, J.A.; Cruz-Oró, E.; Cuéllar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of Flowering and Storage Organ Formation in Potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef]
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science 2010, 328, 872–875. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zheng, Y.; Ham, B.; Zhang, S.; Fei, Z.; Lucas, W.J. Plant LncRNAs Are Enriched in and Move Systemically through the Phloem in Response to Phosphate Deficiency. J. Integr. Plant Biol. 2019, 61, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Kragler, F. Long Distance RNA Movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommer, U.-A.; Thiele, B.-J. The Translationally Controlled Tumour Protein (TCTP). Int. J. Biochem. Cell Biol. 2004, 36, 379–385. [Google Scholar] [CrossRef]
- Hinojosa-Moya, J.J.; Xoconostle-Cázares, B.; Toscano-Morales, R.; Ramírez-Ortega, F.; Luis Cabrera-Ponce, J.; Ruiz-Medrano, R. Characterization of the Pumpkin Translationally-Controlled Tumor Protein CmTCTP. Plant Signal. Behav. 2013, 8, e26477. [Google Scholar] [CrossRef] [Green Version]
- Brioudes, F.; Thierry, A.-M.; Chambrier, P.; Mollereau, B.; Bendahmane, M. Translationally Controlled Tumor Protein Is a Conserved Mitotic Growth Integrator in Animals and Plants. Proc. Natl. Acad. Sci. USA 2010, 107, 16384–16389. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Peng, H.-W.; Cheng, Y.-S.; Yuan, H.S.; Yang-Yen, H.-F. Stabilization and Enhancement of the Antiapoptotic Activity of Mcl-1 by TCTP. Mol. Cell. Biol. 2005, 25, 3117–3126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yang, F.; Xiong, Z.; Yan, Y.; Wang, X.; Nishino, M.; Mirkovic, D.; Nguyen, J.; Wang, H.; Yang, X.-F. An N-Terminal Region of Translationally Controlled Tumor Protein Is Required for Its Antiapoptotic Activity. Oncogene 2005, 24, 4778–4788. [Google Scholar] [CrossRef] [Green Version]
- Rinnerthaler, M.; Jarolim, S.; Heeren, G.; Palle, E.; Perju, S.; Klinger, H.; Bogengruber, E.; Madeo, F.; Braun, R.J.; Breitenbach-Koller, L.; et al. MMI1 (YKL056c, TMA19), the Yeast Orthologue of the Translationally Controlled Tumor Protein (TCTP) Has Apoptotic Functions and Interacts with Both Microtubules and Mitochondria. Biochim. Biophys. Acta (BBA)-Bioenerg. 2006, 1757, 631–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, C.H.; Poon, M.W.; Lun, H.M.; Kwok, P.Y.; Ko, R.C. Heat-Inducible Translationally Controlled Tumor Protein of Trichinella Pseudospiralis: Cloning and Regulation of Gene Expression. Parasitol. Res. 2007, 100, 1105–1111. [Google Scholar] [CrossRef]
- Betsch, L.; Boltz, V.; Brioudes, F.; Pontier, G.; Girard, V.; Savarin, J.; Wipperman, B.; Chambrier, P.; Tissot, N.; Benhamed, M.; et al. TCTP and CSN4 Control Cell Cycle Progression and Development by Regulating CULLIN1 Neddylation in Plants and Animals. PLoS Genet 2019, 15, e1007899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.-C.; Chern, J.J.; Cai, Y.; Liu, M.; Choi, K.-W. Drosophila TCTP Is Essential for Growth and Proliferation through Regulation of DRheb GTPase. Nature 2007, 445, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Betsch, L.; Savarin, J.; Bendahmane, M.; Szecsi, J. Roles of the Translationally Controlled Tumor Protein (TCTP) in Plant Development. In TCTP/tpt1-Remodeling Signaling from Stem Cell to Disease; Telerman, A., Amson, R., Eds.; Results and Problems in Cell Differentiation; Springer International Publishing: Cham, Switzerland, 2017; Volume 64, pp. 149–172. ISBN 978-3-319-67590-9. [Google Scholar]
- Kim, Y.-M.; Han, Y.-J.; Hwang, O.-J.; Lee, S.-S.; Shin, A.-Y.; Kim, S.Y.; Kim, J.-I. Overexpression of Arabidopsis Translationally Controlled Tumor Protein Gene AtTCTP Enhances Drought Tolerance with Rapid ABA-Induced Stomatal Closure. Mol. Cells 2012, 33, 617–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Carvalho, M.; Acencio, M.L.; Laitz, A.V.N.; De Araújo, L.M.; De Lara Campos Arcuri, M.; Do Nascimento, L.C.; Maia, I.G. Impacts of the Overexpression of a Tomato Translationally Controlled Tumor Protein (TCTP) in Tobacco Revealed by Phenotypic and Transcriptomic Analysis. Plant Cell Rep. 2017, 36, 887–900. [Google Scholar] [CrossRef] [Green Version]
- Berkowitz, O.; Jost, R.; Pollmann, S.; Masle, J. Characterization of TCTP, the Translationally Controlled Tumor Protein, from Arabidopsis thaliana. Plant Cell 2009, 20, 3430–3447. [Google Scholar] [CrossRef] [Green Version]
- Liao, F.; Wang, L.; Yang, L.-B.; Peng, X.; Sun, M. NtGNL1 Plays an Essential Role in Pollen Tube Tip Growth and Orientation Likely via Regulation of Post-Golgi Trafficking. PLoS ONE 2010, 5, e13401. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Li, L.; Liao, F.; Chen, W. ITRAQ-Based Quantitative Proteomic Analysis Reveals NtGNL1-Dependent Regulatory Network Underlying Endosome Trafficking for Pollen Tube Polar Growth. Plant Physiol. Biochem. 2021, 161, 200–209. [Google Scholar] [CrossRef]
- Toscano-Morales, R.; Xoconostle-Cázares, B.; Martínez-Navarro, A.C.; Ruiz-Medrano, R. AtTCTP2 MRNA and Protein Movement Correlates with Formation of Adventitious Roots in Tobacco. Plant Signal. Behav. 2016, 11, e1071003. [Google Scholar] [CrossRef] [Green Version]
- Branco, R.; Masle, J. Systemic Signalling through Translationally Controlled Tumour Protein Controls Lateral Root Formation in Arabidopsis. J. Exp. Bot. 2019, 70, 3927–3940. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Ge, F. Current Understanding of the TCTP Interactome. In TCTP/tpt1-Remodeling Signaling from Stem Cell to Disease; Telerman, A., Amson, R., Eds.; Results and Problems in Cell Differentiation; Springer International Publishing: Cham, Switzerland, 2017; Volume 64, pp. 127–136. ISBN 978-3-319-67590-9. [Google Scholar]
- Xoconostle-Cázares, B.; Ruiz-Medrano, R. Structure-Function Relationship of TCTP. In TCTP/tpt1-Remodeling Signaling from Stem Cell to Disease; Telerman, A., Amson, R., Eds.; Results and Problems in Cell Differentiation; Springer International Publishing: Cham, Switzerland, 2017; Volume 64, pp. 47–68. ISBN 978-3-319-67590-9. [Google Scholar]
- Toscano-Morales, R.; Xoconostle-Cázares, B.; Martínez-Navarro, A.C.; Ruiz-Medrano, R. Long Distance Movement of an Arabidopsis Translationally Controlled Tumor Protein (AtTCTP2) mRNA and Protein in Tobacco. Front. Plant Sci. 2014, 5, 705. [Google Scholar] [CrossRef] [Green Version]
- Toscano-Morales, R.; Xoconostle-Cázares, B.; Cabrera-Ponce, J.L.; Hinojosa-Moya, J.; Ruiz-Salas, J.L.; Galván-Gordillo, S.V.; Guevara-González, R.G.; Ruiz-Medrano, R. AtTCTP2, an Arabidopsis thaliana Homolog of Translationally Controlled Tumor Protein, Enhances in Vitro Plant Regeneration. Front. Plant Sci. 2015, 6, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giavalisco, P.; Kapitza, K.; Kolasa, A.; Buhtz, A.; Kehr, J. Towards the Proteome OfBrassica Napus Phloem Sap. Proteomics 2006, 6, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Pérez, B.; Ramírez-Pool, J.A.; Núñez-Muñoz, L.A.; Vargas-Hernández, B.Y.; Camacho-Romero, A.; Lara-Villamar, M.; Jiménez-López, D.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. Engineering Macromolecular Trafficking Into the Citrus Vasculature. Front. Plant Sci. 2022, 13, 818046. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Perrera, V.; Saplaoura, E.; Apelt, F.; Bahin, M.; Kramdi, A.; Olas, J.; Mueller-Roeber, B.; Sokolowska, E.; Zhang, W.; et al. M5C Methylation Guides Systemic Transport of Messenger RNA over Graft Junctions in Plants. Curr. Biol. 2019, 29, 2465–2476.e5. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Galeano, D.F.; Toscano-Morales, R.; Calderón-Pérez, B.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. Structural Divergence of Plant TCTPs. Front. Plant Sci. 2014, 5, 361. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Falcón, M.; Bou, J.; Prat, S. Seasonal Control of Tuberization in Potato: Conserved Elements with the Flowering Response. Annu. Rev. Plant Biol. 2006, 57, 151–180. [Google Scholar] [CrossRef]
- Sonnewald, S.; Sonnewald, U. Regulation of Potato Tuber Sprouting. Planta 2014, 239, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Lin, T.; Hannapel, D.J. Targets of the StBEL5 Transcription Factor Include the FT Ortholog StSP6A. Plant Physiol. 2016, 170, 310–324. [Google Scholar] [CrossRef] [Green Version]
- Jing, S.; Jiang, P.; Sun, X.; Yu, L.; Wang, E.; Qin, J.; Zhang, F.; Prat, S.; Song, B. Long-Distance Control of Potato Storage Organ Formation by SELF PRUNING 3D and FLOWERING LOCUS T-like 1. Plant Commun. 2023, 4, 100547. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Chatterjee, M.; Yu, Y.; Suh, S.-G.; Miller, W.A.; Hannapel, D.J. Dynamics of a Mobile RNA of Potato Involved in a Long-Distance Signaling Pathway. Plant Cell 2007, 18, 3443–3457. [Google Scholar] [CrossRef] [Green Version]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A Potential Graft-Transmissible MicroRNA That Modulates Plant Architecture and Tuberization in Solanum Tuberosum Ssp. Andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondhare, K.R.; Natarajan, B.; Banerjee, A.K. Molecular Signals That Govern Tuber Development in Potato. Int. J. Dev. Biol. 2020, 64, 133–140. [Google Scholar] [CrossRef]
- Weeda, S.M.; Mohan Kumar, G.N.; Richard Knowles, N. Developmentally Linked Changes in Proteases and Protease Inhibitors Suggest a Role for Potato Multicystatin in Regulating Protein Content of Potato Tubers. Planta 2009, 230, 73–84. [Google Scholar] [CrossRef]
- Mahajan, A.; Bhogale, S.; Kang, I.H.; Hannapel, D.J.; Banerjee, A.K. The MRNA of a Knotted1-like Transcription Factor of Potato Is Phloem Mobile. Plant Mol. Biol. 2012, 79, 595–608. [Google Scholar] [CrossRef]
- Cho, S.K.; Sharma, P.; Butler, N.M.; Kang, I.-H.; Shah, S.; Rao, A.G.; Hannapel, D.J. Polypyrimidine Tract-Binding Proteins of Potato Mediate Tuberization through an Interaction with StBEL5 RNA. EXBOTJ 2015, 66, 6835–6847. [Google Scholar] [CrossRef] [Green Version]
- Kondhare, K.R.; Kumar, A.; Hannapel, D.J.; Banerjee, A.K. Conservation of Polypyrimidine Tract Binding Proteins and Their Putative Target RNAs in Several Storage Root Crops. BMC Genom. 2018, 19, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naing, A.H.; Kim, C.K. Abiotic Stress-induced Anthocyanins in Plants: Their Role in Tolerance to Abiotic Stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Teo, C.-J.; Takahashi, K.; Shimizu, K.; Shimamoto, K.; Taoka, K. Potato Tuber Induction Is Regulated by Interactions Between Components of a Tuberigen Complex. Plant Cell Physiol. 2017, 58, 365–374. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Lee, D.-H. Use of Serial Analysis of Gene Expression Technology to Reveal Changes in Gene Expression in Arabidopsis Pollen Undergoing Cold Stress. Plant Physiol. 2003, 132, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Sage-Ono, K.; Ono, M.; Harada, H.; Kamada, H. Dark-Induced Accumulation of MRNA for a Homolog of Translationally Controlled Tumor Protein (TCTP) in Pharbitis. Plant Cell Physiol. 1998, 39, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Kumlay, A.M.; Ercisli, S. Callus Induction, Shoot Proliferation and Root Regeneration of Potato (Solanum tuberosum L.) Stem Node and Leaf Explants under Long-Day Conditions. Biotechnol. Biotechnol. Equip. 2015, 29, 1075–1084. [Google Scholar] [CrossRef] [Green Version]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved Method for the Isolation of RNA from Plant Tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Muñoz, L.; Vargas-Hernández, B.; Hinojosa-Moya, J.; Ruiz-Medrano, R.; Xoconostle-Cázares, B. Plant Drought Tolerance Provided through Genome Editing of the Trehalase Gene. Plant Signal. Behav. 2021, 16, 1877005. [Google Scholar] [CrossRef] [PubMed]
- Mba’u, Y.J.; Iriawati; Faizal, A. Transient Transformation of Potato Plant (Solanum tuberosum L.) Granola Cultivar Using Syringe Agroinfiltration. Agrivita. J. Agr. Sci. 2018, 40, 1456. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Salas, J.-L.; Ruiz-Medrano, R.; Montes-Horcasitas, M.D.C.; Agreda-Laguna, K.A.; Hinojosa-Moya, J.; Xoconostle-Cázares, B. Vascular Expression of Trehalose Phosphate Synthase1 (TPS1) Induces Flowering in Arabidopsis. Plant Omics 2016, 9, 344–351. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hofacker, I.L. Vienna RNA Secondary Structure Server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xiong, Y.; Xiao, Y. 3dDNA: A Computational Method of Building DNA 3D Structures. Molecules 2022, 27, 5936. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Xiao, Y. 3dRNA: 3D Structure Prediction from Linear to Circular RNAs. J. Mol. Biol. 2022, 434, 167452. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation: PROTEIN SCIENCE.ORG. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Wadley, L.M.; Keating, K.S.; Duarte, C.M.; Pyle, A.M. Evaluating and Learning from RNA Pseudotorsional Space: Quantitative Validation of a Reduced Representation for RNA Structure. J. Mol. Biol. 2007, 372, 942–957. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Jumper, J.; Hassabis, D. Protein Structure Predictions to Atomic Accuracy with AlphaFold. Nat Methods 2022, 19, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Shuvo, M.H.; Gulfam, M.; Bhattacharya, D. DeepRefiner: High-Accuracy Protein Structure Refinement by Deep Network Calibration. Nucleic Acids Res. 2021, 49, W147–W152. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A Web Server for Protein–Protein and Protein–DNA/RNA Docking Based on a Hybrid Strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK Server for Integrated Protein–Protein Docking. Nat Protoc 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xoconostle-Morán, B.B.; Xoconostle-Cázares, B.; Vargas-Hernández, B.Y.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; Ruiz-Medrano, R. Long-Distance Movement of Solanum tuberosum Translationally Controlled Tumor Protein (StTCTP) mRNA. Plants 2023, 12, 2839. https://doi.org/10.3390/plants12152839
Xoconostle-Morán BB, Xoconostle-Cázares B, Vargas-Hernández BY, Núñez-Muñoz LA, Calderón-Pérez B, Ruiz-Medrano R. Long-Distance Movement of Solanum tuberosum Translationally Controlled Tumor Protein (StTCTP) mRNA. Plants. 2023; 12(15):2839. https://doi.org/10.3390/plants12152839
Chicago/Turabian StyleXoconostle-Morán, Brenda Beatriz, Beatriz Xoconostle-Cázares, Brenda Yazmín Vargas-Hernández, Leandro Alberto Núñez-Muñoz, Berenice Calderón-Pérez, and Roberto Ruiz-Medrano. 2023. "Long-Distance Movement of Solanum tuberosum Translationally Controlled Tumor Protein (StTCTP) mRNA" Plants 12, no. 15: 2839. https://doi.org/10.3390/plants12152839





