Structural and Functional Analysis of the MADS-Box Genes Reveals Their Functions in Cold Stress Responses and Flower Development in Tea Plant (Camellia sinensis)
Abstract
1. Introduction
2. Results
2.1. Identification of the MADS-Box Gene Family Based on Tea-Specific Nucleic Acid HMM and the Curation of Gene Models
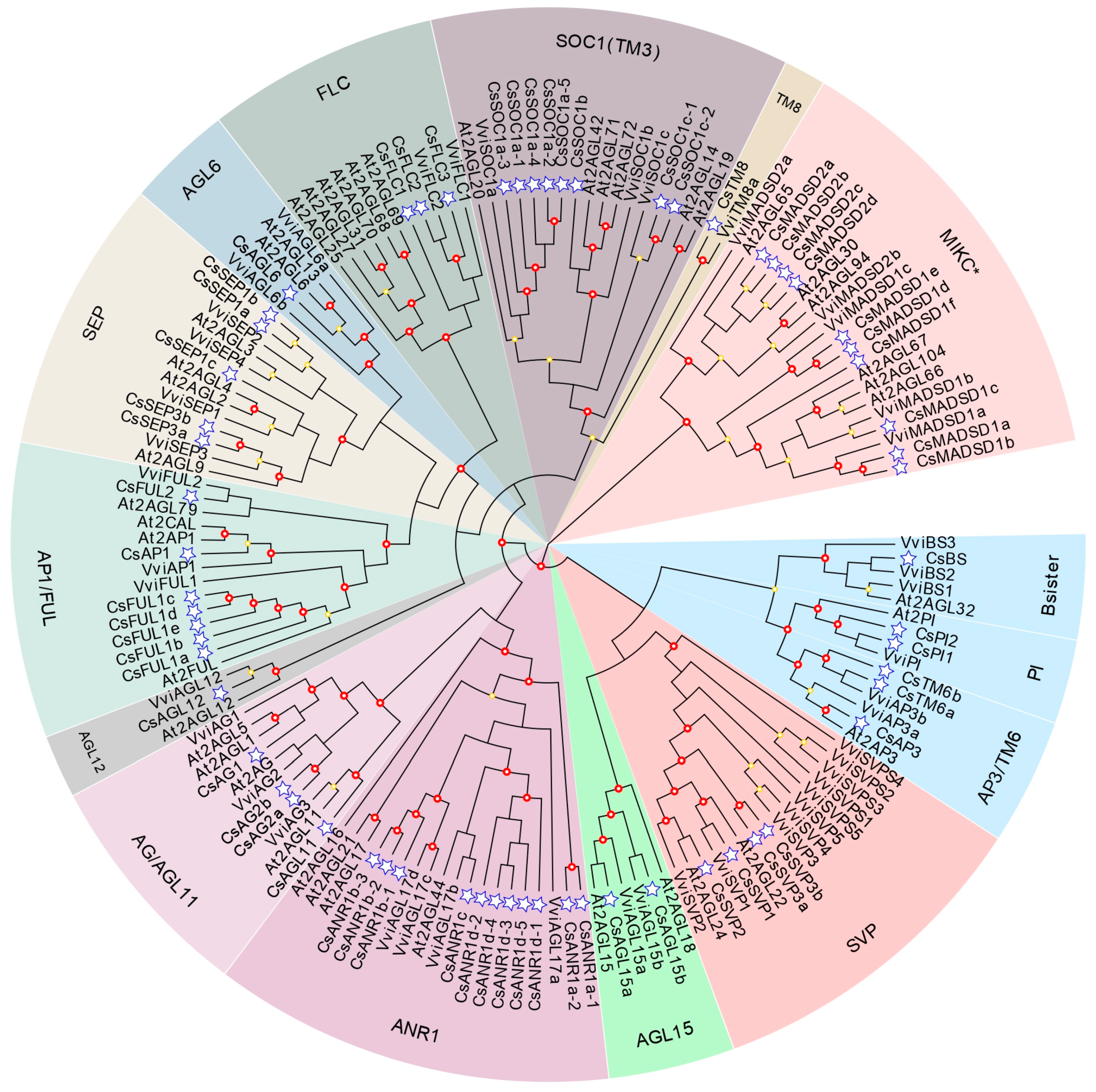
2.2. Maximum Likelihood Phylogenetic Analysis of MADS-Box Genes
2.3. Analysis of Genes Structure and Conserved Motifs
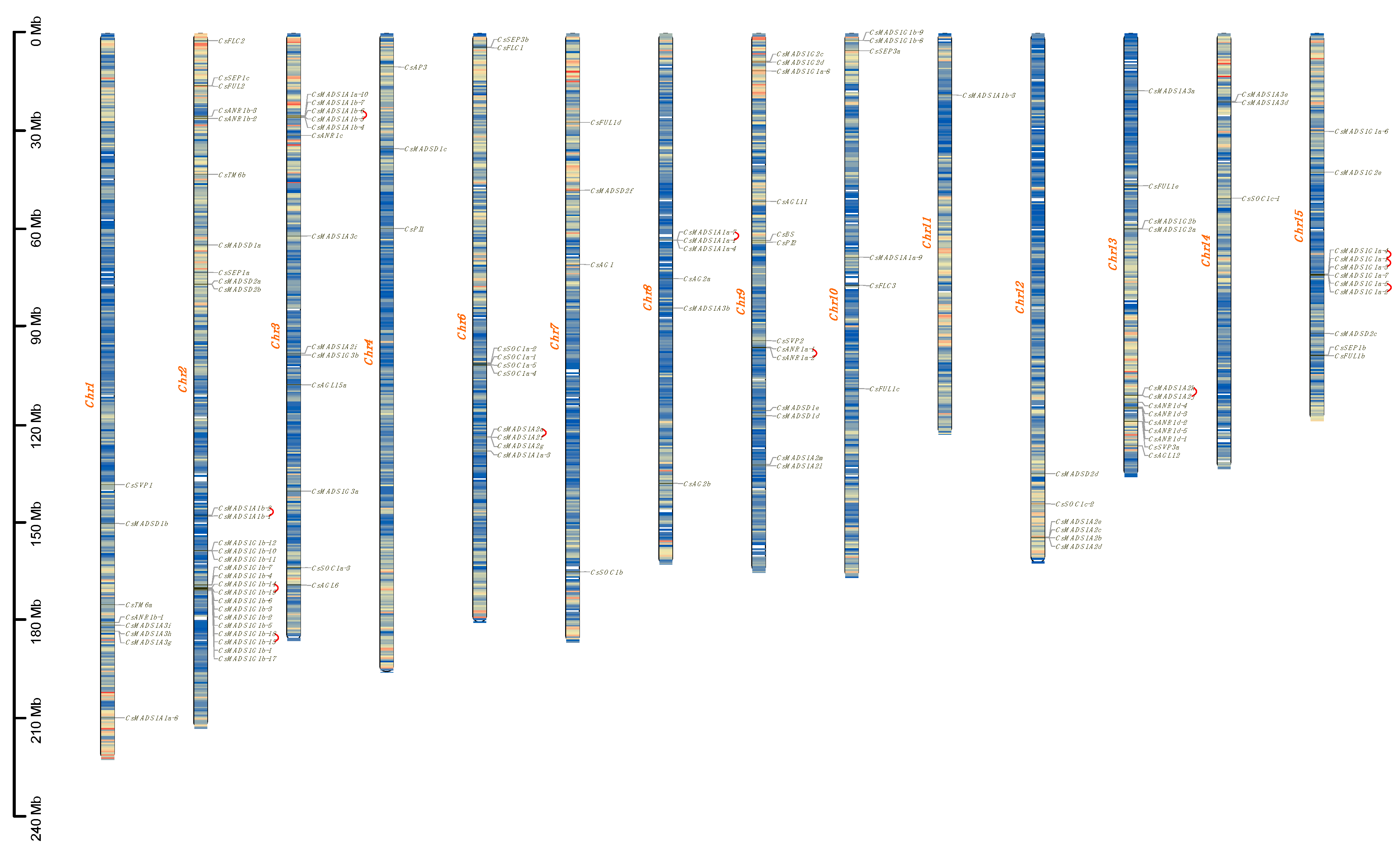
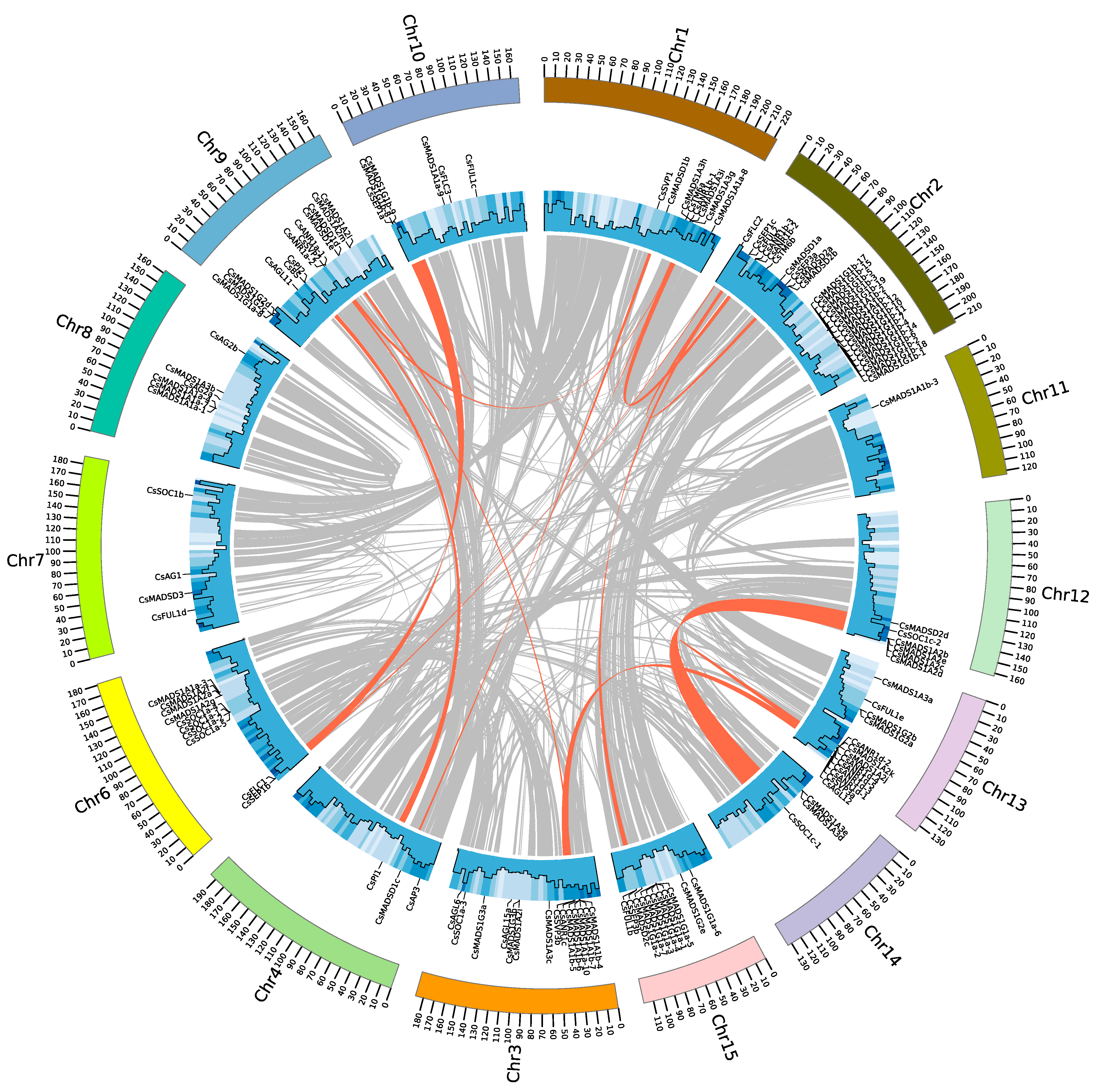
2.4. Chromosomal Location and Gene Duplication Events of the MADS-Box Gene Family in Tea Plant
2.5. MADS-Box Gene Expression Analysis among Different Tissues and Seasons
2.6. MADS-Box Gene Expression Analysis in Response to Cold Stress
2.7. A Putative Floral ABCE Model for Tea Plant According to Expression Profiles
3. Discussion
3.1. Evolution of the MADS-Box Family in Tea Plant
3.2. Extensively Conserved Function of MADS-Box Genes in Tea Plant Development
3.3. The Involvement of Tea Plant MADS-Box Genes in Cold Stress Responses
3.4. A Conserved and Ancient Floral ‘ABCE’ Model in Tea Plant
4. Materials and Methods
4.1. Identification of MADS-Box Genes and Curation of a Gene Model
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Gene Nomenclature of Tea Plant MADS-Box Genes
4.4. Characterization of the Gene Structure and Conserved Motifs of CsMADS
4.5. Gene Mapping, Gene Duplication, Synteny, and Ka/Ks Substitutions of MADS-Box Genes in Tea Plant
4.6. Gene Expression Analysis of Tea Plant MADS-Box Genes
4.7. Plant Materials and Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fan, Z.; Li, J.; Li, X.; Wu, B.; Wang, J.; Liu, Z.; Yin, H. Genome-wide transcriptome profiling provides insights into floral bud development of summer-flowering Camellia azalea. Sci. Rep. 2015, 5, 9729. [Google Scholar] [CrossRef]
- Pereira, A.G.; Garcia-Perez, P.; Cassani, L.; Chamorro, F.; Cao, H.; Barba, F.J.; Simal-Gandara, J.; Prieto, M.A. Camellia japonica: A phytochemical perspective and current applications facing its industrial exploitation. Food Chem. X 2022, 13, 100258. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Li, W.; Li, K.; Nan, H.; Shi, C.; Zhang, Y.; Dai, Z.Y.; Lin, Y.L.; Yang, X.L.; Tong, Y.; et al. The Chromosome-Level Reference Genome of Tea Tree Unveils Recent Bursts of Non-autonomous LTR Retrotransposons in Driving Genome Size Evolution. Mol. Plant 2020, 13, 935–938. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.; Qiu, H.; Guo, Y.; Wan, H.; Zhang, X.; Scossa, F.; Alseekh, S.; Zhang, Q.; Wang, P.; et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 2020, 11, 3719. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef] [PubMed]
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B.K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Martinez-Castilla, L.P.; Alvarez-Buylla, E.R. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc. Natl. Acad. Sci. USA 2003, 100, 13407–13412. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martinez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef]
- Kofuji, R.; Sumikawa, N.; Yamasaki, M.; Kondo, K.; Ueda, K.; Ito, M.; Hasebe, M. Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 2003, 20, 1963–1977. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Becker, A. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Smyth, D.R. Evolution and genetic control of the floral ground plan. New Phytol. 2018, 220, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. The ABC model of flower development: Then and now. Development 2012, 139, 4095–4098. [Google Scholar] [CrossRef]
- Jofuku, K.D.; den Boer, B.G.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar]
- Mandel, M.A.; Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Savidge, B.; Rounsley, S.D.; Yanofsky, M.F. Temporal Relationship between the Transcription of Two Arabidopsis MADS-Box Genes and the Floral Organ ldentity Genes. Plant Cell 1995, 7, 721–733. [Google Scholar]
- Pelaz, S.; Ditta, G.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C foral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Hsu, H.-F.; Hsu, W.-H.; Lee, Y.-I.; Mao, W.-T.; Yang, J.-Y.; Li, J.-Y.; Yang, C.-H. Model for perianth formation in orchids. Nat. Plants 2015, 1, 15046. [Google Scholar] [CrossRef]
- Suetsugu, K.; Fukushima, K.; Makino, T.; Ikematsu, S.; Sakamoto, T.; Kimura, S. Transcriptomic heterochrony and completely cleistogamous flower development in the mycoheterotrophic orchid Gastrodia. New Phytol. 2023, 237, 323–338. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-Box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef]
- Lozano, R.; Angosto, T.; Gomez, P.; Payan, C.; Capel, J.; Huijser, P.; Salinas, J.; Martinez-Zapater, J.M. Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-Box genes. Plant Physiol. 1998, 117, 91–100. [Google Scholar] [CrossRef]
- Bartley, G.E.; Ishida, B.K. Developmental gene regulation during tomato fruit ripening and in-vitro sepal morphogenesis. BMC Plant Biol. 2003, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Jin, Y.J.; Wan, H.H.; Cheng, L.; Feng, Z.G. Genome-wide identification and expression analysis of the MADS-box transcription factor family in Camellia sinensis. J. Appl. Genet. 2021, 62, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Liu, X.; Vanneste, K.; Proost, S.; Tsai, W.C.; Liu, K.W.; Chen, L.J.; He, Y.; Xu, Q.; Bian, C.; et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef]
- Lu, H.; Liu, Z.; Lan, S. Genome Sequencing Reveals the Role of MADS-box Gene Families in the Floral Morphology Evolution of Orchids. Hortic. Plant J. 2019, 5, 247–254. [Google Scholar] [CrossRef]
- Cheng, Z.; Ge, W.; Li, L.; Hou, D.; Ma, Y.; Liu, J.; Bai, Q.; Li, X.; Mu, S.; Gao, J. Analysis of MADS-Box Gene Family Reveals Conservation in Floral Organ ABCDE Model of Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 2017, 8, 656. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, D.; Lin, X.; Ding, M.; Tong, Z. Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol. 2018, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Zhao, T.; Holmer, R.; de Bruijn, S.; Angenent, G.C.; van den Burg, H.A.; Schranz, M.E. Phylogenomic Synteny Network Analysis of MADS-Box Transcription Factor Genes Reveals Lineage-Specific Transpositions, Ancient Tandem Duplications, and Deep Positional Conservation. Plant Cell 2017, 29, 1278–1292. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Z.; Li, S.; Hou, M.; Zhou, Y.; Zhao, Y.; Li, G.; Zhao, H.; Ma, H. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef]
- Wells, C.E.; Vendramin, E.; Jimenez Tarodo, S.; Verde, I.; Bielenberg, D.G. A genome-wide analysis of MADS-box genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2015, 15, 41. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, A.; Rajput, R.; Sidhu, S.; Dhillon, H.; Verma, P.C.; Pandey, A.; Upadhyay, S.K. Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals. Plants 2022, 11, 911. [Google Scholar] [CrossRef]
- Rathour, M.; Shumayla; Alok, A.; Upadhyay, S.K. Investigation of Roles of TaTALE Genes during Development and Stress Response in Bread Wheat. Plants 2022, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Heijmans, K.; Airoldi, C.; Davies, B.; Angenent, G.C. An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 2010, 154, 287–300. [Google Scholar] [CrossRef]
- Gramzow, L.; Theissen, G. Phylogenomics of MADS-Box Genes in Plants-Two Opposing Life Styles in One Gene Family. Biology 2013, 2, 1150–1164. [Google Scholar] [CrossRef] [PubMed]
- Grimplet, J.; Martinez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Kramer, E.M.; Su, H.J.; Wu, C.C.; Hu, J.M. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evol. Biol. 2006, 6, 30. [Google Scholar] [CrossRef]
- Airoldi, C.A.; Davies, B. Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genom. 2012, 39, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Lopez, R.; Garcia-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Perez-Ruiz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef]
- Diaz-Riquelme, J.; Lijavetzky, D.; Martinez-Zapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhao, P.; Cheng, L.; Yuan, G.; Yang, W.; Liu, S.; Chen, S.; Qi, D.; Liu, G.; Li, X. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 2018, 18, 42. [Google Scholar] [CrossRef]
- Shimada, S.; Ogawa, T.; Kitagawa, S.; Suzuki, T.; Ikari, C.; Shitsukawa, N.; Abe, T.; Kawahigashi, H.; Kikuchi, R.; Handa, H.; et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 2009, 58, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Hemming, M.N.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008, 147, 355–366. [Google Scholar] [CrossRef]
- Ishida, B.K.; Jenkins, S.M.; Say, B. Induction of AGAMOUS gene expression plays a key role in ripening of tomato sepals in vitro. Plant Mol. Biol. 1998, 36, 733–739. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Seo, E.; Lee, H.; Jeon, J.; Park, H.; Kim, J.; Noh, Y.-S.; Lee, I. Crosstalk between Cold Response and Flowering in Arabidopsis Is Mediated through the Flowering-Time Gene SOC1 and Its Upstream Negative Regulator FLC. Plant Cell 2009, 21, 3185–3197. [Google Scholar] [CrossRef] [PubMed]
- Sarah, J.; Gilmour, S.G.F.a.M.F.T. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar]
- Wu, R.M.; Walton, E.F.; Richardson, A.C.; Wood, M.; Hellens, R.P.; Varkonyi-Gasic, E. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 2012, 63, 797–807. [Google Scholar] [CrossRef]
- Liu, X.; Sun, Z.; Dong, W.; Wang, Z.; Zhang, L. Expansion and Functional Divergence of the SHORT VEGETATIVE PHASE (SVP) Genes in Eudicots. Genome Biol. Evol. 2018, 10, 3026–3037. [Google Scholar] [CrossRef]
- Yamane, H.; Ooka, T.; Jotatsu, H.; Hosaka, Y.; Sasaki, R.; Tao, R. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 2011, 62, 3481–3488. [Google Scholar] [CrossRef]
- Litt, A.; Kramer, E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 2010, 21, 129–137. [Google Scholar] [CrossRef]
- Sharma, B.; Kramer, E.M. Aquilegia B gene homologs promote petaloidy of the sepals and maintenance of the C domain boundary. Evodevo 2017, 8, 22. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; Yoo, M.J.; Zahn, L.M.; Brockington, S.F.; Wall, P.K.; Gitzendanner, M.A.; Albert, V.A.; Leebens-Mack, J.; Altman, N.S.; Ma, H.; et al. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc. Natl. Acad. Sci. USA 2010, 107, 22570–22575. [Google Scholar] [CrossRef]
- Gaopu, Z. The potential role of B-function gene involved in floral development for double flowers formation in Camellia changii Ye. Afr. J. Biotechnol. 2011, 10, 541–548. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Fan, Z.; Liu, Z.; Tanaka, T.; Yin, H. Global gene expression defines faded whorl specification of double flower domestication in Camellia. Sci. Rep. 2017, 7, 3197. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Li, F.D.; Tong, W.; Li, P.H.; Wu, Q.; Zhao, H.J.; Ge, R.H.; Li, R.P.; Li, Y.Y.; Zhang, Z.Z.; et al. Tea Plant Information Archive: A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 2019, 17, 1938–1953. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Yue, C.; Chen, Q.; Hu, J.; Li, C.; Luo, L.; Zeng, L. Genome-Wide Identification and Characterization of GARP Transcription Factor Gene Family Members Reveal Their Diverse Functions in Tea Plant (Camellia sinensis). Front. Plant Sci. 2022, 13, 947072. [Google Scholar] [CrossRef]
- Hao, X.; Tang, H.; Wang, B.; Yue, C.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Integrative transcriptional and metabolic analyses provide insights into cold spell response mechanisms in young shoots of the tea plant. Tree Physiol. 2018, 38, 1655–1671. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Ban, Q.; Zhu, X.; Jiang, C.; Wei, C.; Bennetzen, J.L. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis. BMC Genom. 2019, 20, 624. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Chen, Q.; Idrees, A.; Bi, W.; Lai, Z.; Sun, Y. Structural and Functional Analysis of the MADS-Box Genes Reveals Their Functions in Cold Stress Responses and Flower Development in Tea Plant (Camellia sinensis). Plants 2023, 12, 2929. https://doi.org/10.3390/plants12162929
Hu J, Chen Q, Idrees A, Bi W, Lai Z, Sun Y. Structural and Functional Analysis of the MADS-Box Genes Reveals Their Functions in Cold Stress Responses and Flower Development in Tea Plant (Camellia sinensis). Plants. 2023; 12(16):2929. https://doi.org/10.3390/plants12162929
Chicago/Turabian StyleHu, Juan, Qianqian Chen, Atif Idrees, Wanjun Bi, Zhongxiong Lai, and Yun Sun. 2023. "Structural and Functional Analysis of the MADS-Box Genes Reveals Their Functions in Cold Stress Responses and Flower Development in Tea Plant (Camellia sinensis)" Plants 12, no. 16: 2929. https://doi.org/10.3390/plants12162929
APA StyleHu, J., Chen, Q., Idrees, A., Bi, W., Lai, Z., & Sun, Y. (2023). Structural and Functional Analysis of the MADS-Box Genes Reveals Their Functions in Cold Stress Responses and Flower Development in Tea Plant (Camellia sinensis). Plants, 12(16), 2929. https://doi.org/10.3390/plants12162929







