Abstract
The infection of soil-borne diseases has the potential to modify root exudation and the rhizosphere microbiome. However, the extent to which these modifications occur in various monocropping histories remains inadequately explored. This study sampled healthy and diseased American ginseng (Panax quinquefolius L.) plants under 1–4 years of monocropping and analyzed the phenolic acids composition by HPLC, microbiome structure by high-throughput sequencing technique, and the abundance of pathogens by quantitative PCR. First, the fungal pathogens of Fusarium solani and Ilyonectria destructans in the rhizosphere soil were more abundant in the diseased plants than the healthy plants. The healthy American ginseng plants exudated more phenolic acid, especially p-coumaric acid, compared to the diseased plants after 1–2 years of monocropping, while this difference gradually diminished with the increase in monocropping years. The pathogen abundance was influenced by the exudation of phenolic acids, e.g., total phenolic acids (r = −0.455), p-coumaric acid (r = −0.465), and salicylic acid (r = −0.417), and the further in vitro test confirmed that increased concentration of p-coumaric acid inhibited the mycelial growth of the isolated pathogens for root rot. The healthy plants had a higher diversity of rhizosphere bacterial and fungal microbiome than the diseased plants only after a long period of monocropping. Our study has revealed that the cropping history of American ginseng has altered the effect of pathogens infection on rhizosphere microbiota and root exudation.
1. Introduction
American ginseng (Panax quinquefolius L.) is one of the most important medical plants in the world and has been widely cultivated under monocropping to increase productivity. However, this practice has also shown negative impacts on American ginseng’s medicinal quality [1]. Monocropping faces various challenges, from the reduction of microbial diversity in the soil [2], degradation of soil organic matter [3], plant autotoxicity [4], and increased susceptibility to soil-borne diseases [5].
A rhizosphere microbial community, including soil-borne diseases, plays an important role in determining the health of Panax plants [6,7,8]. The pathogens for American ginseng root rot include Fusarium solani, F. oxysporum, and Cylindrocarpon destructans [1,9], which can damage the seedling’s roots, and mainly spread through soil moisture, underground insects, and nematodes [10]. The previous study on healthy and diseased P. notoginseng showed that the differed composition of bacterial and fungal communities in the rhizosphere soil [7] and the abundance of Pseudomonas spp. and Ilyonectria spp. in the diseased plants were significantly higher than the healthy plants, confirming the interactions between pathogen diseases and the microbiome [11]. On the other hand, compared with the healthy citrus, the relative abundances of the potentially plant-beneficial bacteria Bradyrhizobium spp. and Burkholderia spp. were lower in the root of the diseased plant [12]. Burkholderia spp. could degrade the fungal cell wall through their hydrolytic and catalytic enzymes [13]. In addition, several ISR (Induced Systemic Resistance)-associated genes in the plant, such as SAM, PR1, and PR2, were induced by Bradyrhizobium spp. and Burkholderia spp. [14].
In the monocropping system, where the same crop is cultivated repeatedly, there is a risk of decreased diversity of microorganisms in the rhizosphere [15]. This reduction may be caused by nutrient depletion in the soil and the accumulation of pathogenic microorganisms [16]. Studies have demonstrated that monocropping can lead to a decline in the abundance and diversity of the plant growth-promoting microbes in the rhizosphere, e.g., Burkholderia spp., Pseudomonas spp., and Trichoderma spp. [17], which play essential roles in improving plant nutrient uptake, disease suppression, and root development [18]. However, there are few studies evaluating the disease effect on the American ginseng rhizosphere microbial communities across various monocropping histories.
Phenolic acids exudated from plant roots can change the structure of soil microorganisms by stimulating soil-borne pathogens and inhibiting beneficial microbes [6,19,20]. For example, wilt-diseased tomato (Solanum lycopersicum) had higher concentrations of p-hydroxybenzoic acid, vanillic acid, and ferulic acid in the rhizosphere soil than the healthy plants, and the increased concentrations of phenolic acids significantly stimulated F. solani growth [21]. The exudation of cinnamic acid increased the incidence rates of Fusarium wilt disease in faba bean (Vicia faba L.) [22]. In the soil planted with P. notoginseng, the total amount of phenolic acids increased with the continuous planting years [11]. For American ginseng, the growth of the pathogen species Rhizoctonia solani was promoted by the low-level exudation of phenolic acids, e.g., p-coumaric acid, syringic acid, and vanillic acid, but suppressed by the higher concentrations of phenolic acid [23]. Phenolic acid in the rhizosphere soil of American ginseng was changed with the increase in monocropping years and drove the dynamics of the pathogenic microorganisms [24]. Therefore, it is crucial to identify the interactions among diverse phenolic acids, microbiome, and pathogens’ occurrence in the rhizosphere microhabitat of American ginseng.
The objectives of this study were to investigate the variations of phenolic acids and microorganisms in the rhizosphere soil of the healthy and diseased American ginseng plants cultivated under continuous monocropping practices and explore the relationships between the phenolic acids, rhizosphere microbiome structure, and pathogen abundance in the rhizosphere.
2. Result
2.1. The Abundance of American Ginseng Pathogens in the Rhizosphere Soil
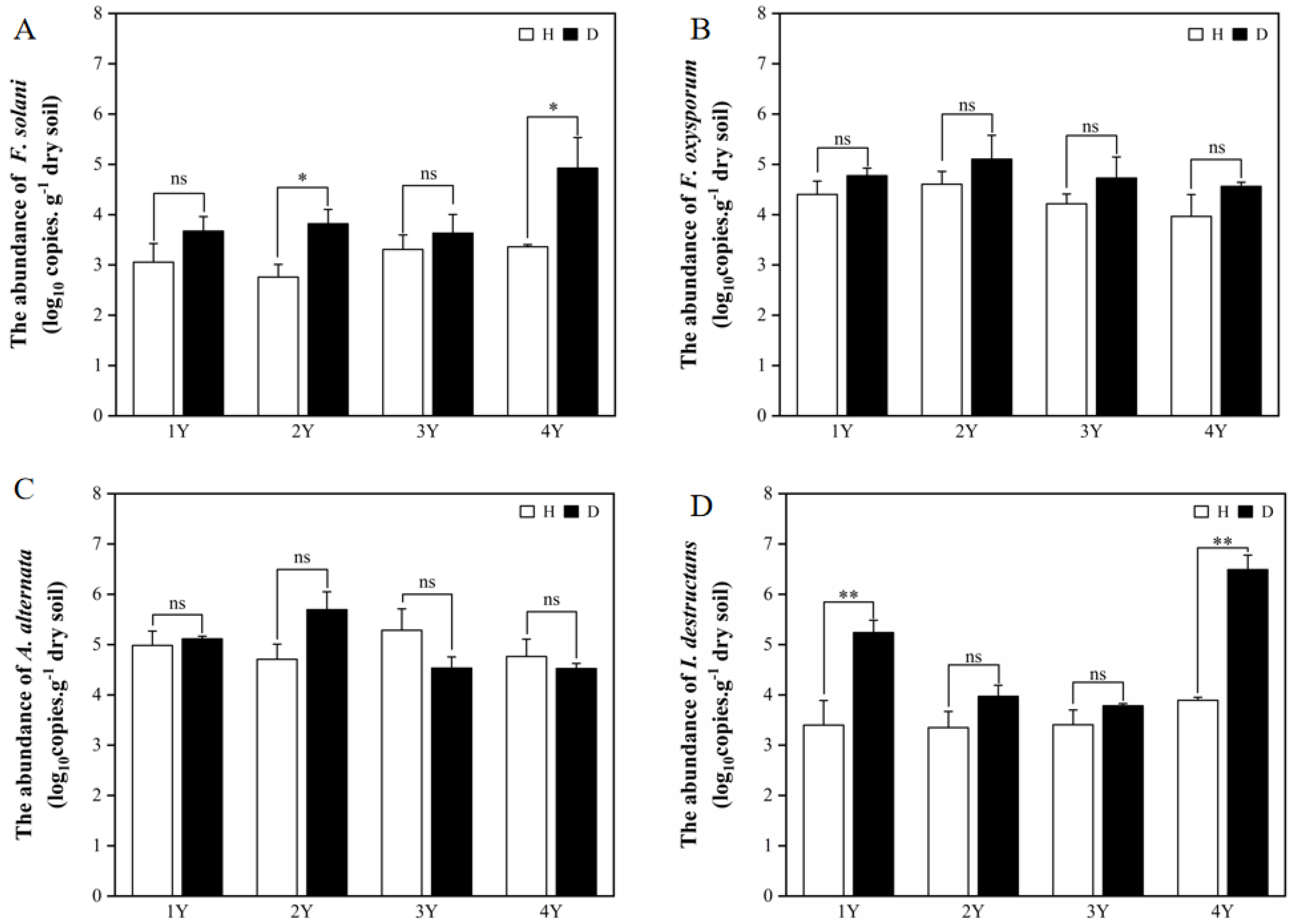
Four common pathogens (F. solani, F. oxysporum, A. alternata, and I. destructans) of American ginseng were selected for the qPCR analysis. The difference in pathogens abundance in the healthy and diseased American ginseng was affected by monocropping years. In general, the presence of pathogens in the rhizosphere soil of the diseased American ginseng was either greater or comparable to that of the healthy plants (Figure 1).
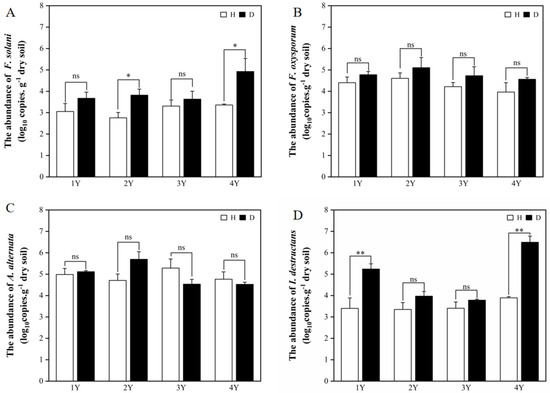
Figure 1.
The abundance of F. solani (A), F. oxysporum (B), A. alternata (C), and I. destructans (D) in the rhizosphere soils of the healthy (H) and diseased (D) American ginseng in 1–4 years of continuous monocropping (1Y, 2Y, 3Y, and 4Y). Error bars indicate the standard errors. ns indicates no significant difference at p < 0.05 according to Duncan’s multiple range test, * and ** indicate significant differences at p < 0.05 and p < 0.01, respectively.
The abundance of F. solani in the diseased American ginseng was significantly higher than that in the healthy plants with under 2 and 4 years of monocropping (Figure 1A). While under 1 and 3 years of monocropping, the disease infection had no influence on the abundance of F. solani in the rhizosphere soil. For the abundance of F. oxysporum and A. alternata in the rhizosphere soil, there was no difference between diseased and healthy American ginseng (Figure 1B,C). Compared with healthy plants, the I. destructans abundance in the rhizosphere soil of diseased American ginseng increased by 30.99% and 36.04% under 1 and 4 years of monocropping history, respectively (Figure 1D), while the difference was not significant under 2 or 3 years of monocropping history.
2.2. Phenolic Acid Content in the Rhizosphere Soil of American Ginseng
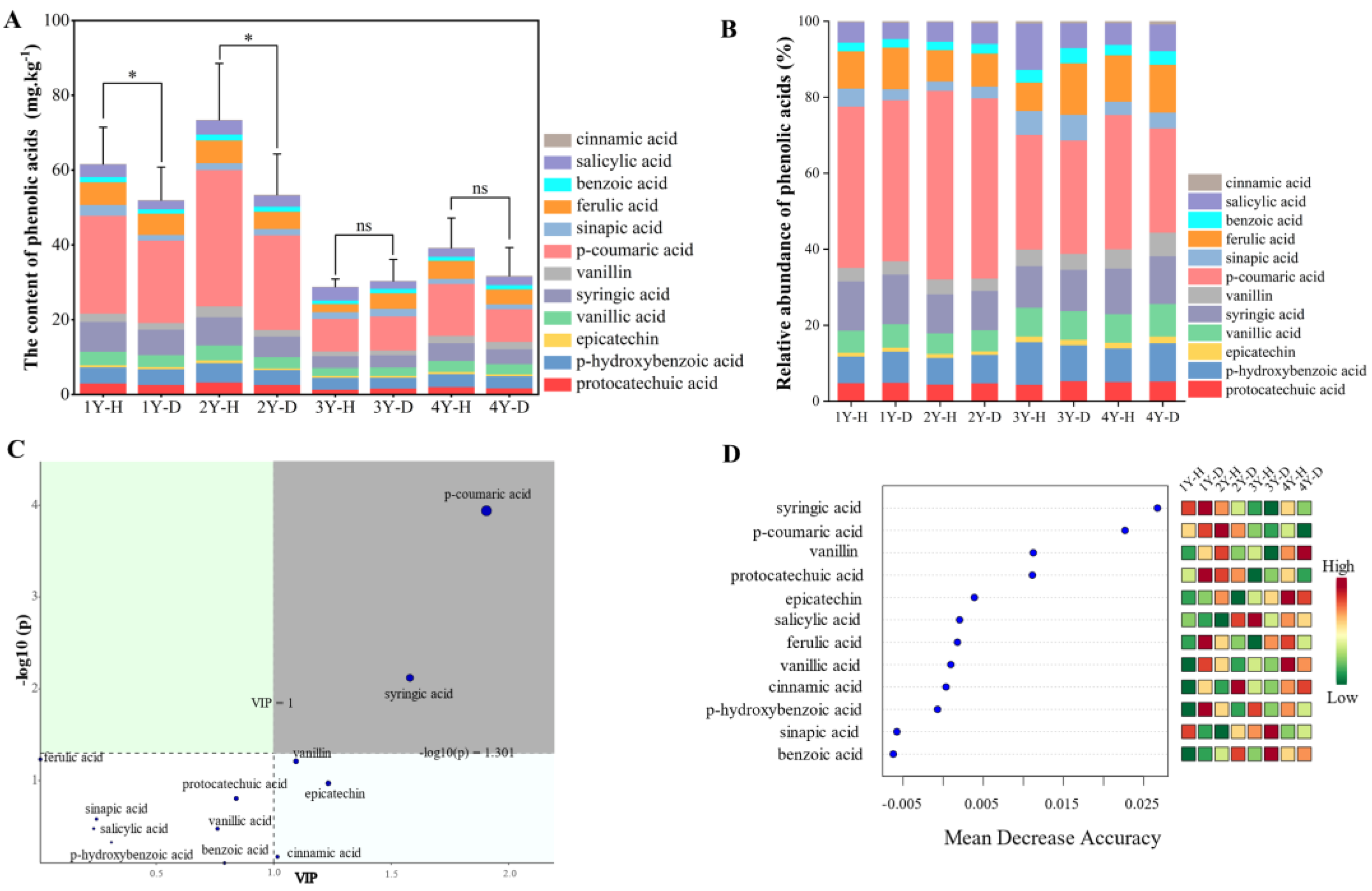
In total, 12 compounds of phenolic acids were detected by the HPLC system according to their retention time as protocatechuic acid, epicatechin, p-hydroxybenzoic acid, vanillic acid, erucic acid, syringic acid, p-coumaric acid, vanillin, ferulic acid, benzoic acid, salicylic acid, and cinnamic acid (Figure 2A). Compared with the healthy plants, the content of total phenolic acids in the rhizosphere soil of the diseased American ginseng significantly reduced by 15.65% and 27.30% under 1 and 2 monocropping years, respectively. The disease effect was not significant when the monocropping history became longer, 3 and 4 years (Figure 2A). The content of phenolic acid in the rhizosphere soil of American ginseng decreased with the increase in monocropping years.
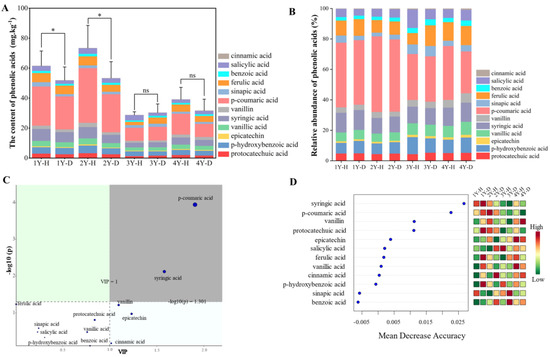
Figure 2.
The phenolic acids content (A) and composition (B), the VIP of PLS-DA (C), and the random forest analysis (D) of phenolic acids in the rhizosphere soil of the healthy (H) and diseased (D) American ginseng in 1–4 years of monocropping (1Y, 2Y, 3Y, and 4Y). Error bars indicate the standard errors. ns indicates no significant difference at p < 0.05 according to Duncan’s multiple range test, * indicate significant differences at p < 0.05.
The p-coumaric acid had the highest relative content among the 12 phenolic acids in the rhizosphere soil of American ginseng, about 40.57% (Figure 2B). The concentration of p-coumaric acid exhibited a gradual decline with increasing years of monocropping, from 42.42% in 1 year of monocropping to 31.84% in 4 years of monocropping. The rhizosphere soil of diseased American ginseng showed a decreased level of p-coumaric acid in comparison to healthy plants (38.88% vs. 41.96%). Likewise, when compared to healthy plants, the benzoic acid content in the rhizosphere soil of American ginseng that had been diseased for 1 year exhibited a significant reduction of 16.30%. However, there were no significant differences in benzoic acid levels between healthy and diseased soil in the 2–4 years of monocropping. The VIP diagram of PLS-DA (Figure 2C) and the random forest analysis (Figure 2D) revealed that p-coumaric acid and syringic acid were the major metabolites driving the variation in phenolic acid composition in the rhizosphere soil of healthy and diseased American ginseng with varied monocropping years (p < 0.05).
2.3. Microbial Communities in the Rhizosphere Soil of the Healthy and Diseased American Ginseng
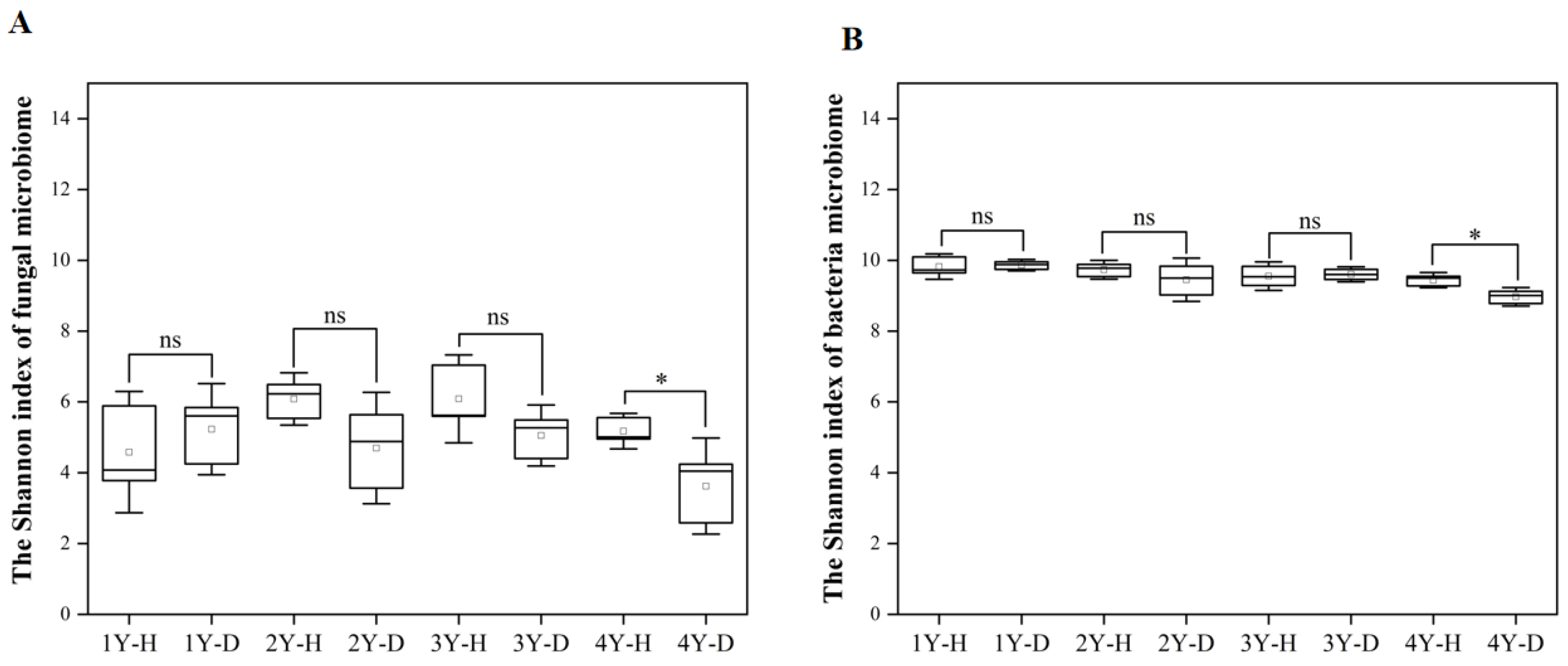
The Shannon index was used to indicate the alpha diversity of rhizosphere soil fungal and bacterial microbiome in each sample (Figure 3A,B). For both fungal and bacterial communities, the Shannon index in the rhizosphere soil of healthy American ginseng was higher in comparison to diseased plants only under 4 years of monocropping (Figure 3).
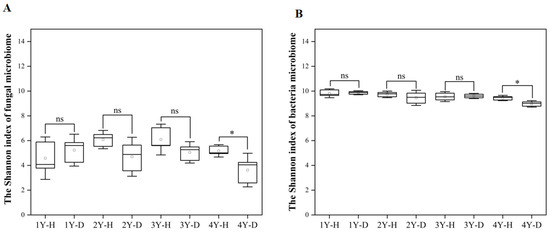
Figure 3.
The Shannon index of fungal (A) and bacterial (B) microbiomes in the rhizosphere soil of the healthy (H) and diseased (D) American ginseng in 1–4 years of continuous monocropping (1Y, 2Y, 3Y, and 4Y). Error bars indicate the standard errors. ns indicates no significant difference at p < 0.05 according to Duncan’s multiple range test, * indicate significant difference sat p < 0.05.
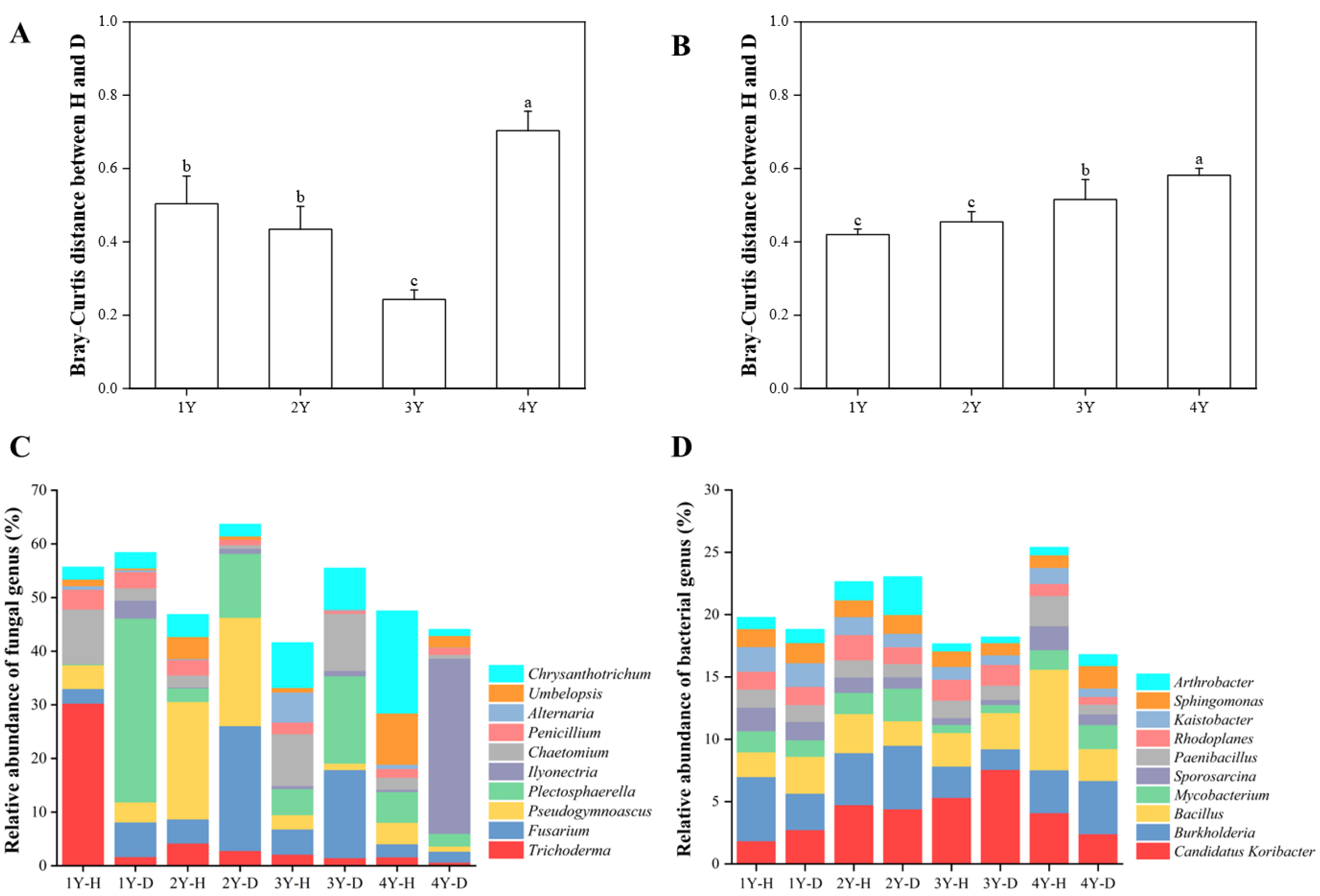
The fungal (Figure S1A) and bacterial (Figure S1B) microbiome in the rhizosphere soil of healthy and diseased American ginseng during monocropping years were compared using PCoA analysis based on the Bray–Curtis distance. The results showed that the first principal coordinates explained 40.60% and 39.70% of the total variation for the fungal and bacterial microbiomes, respectively. The fungal and bacterial microbiomes in diseased soil after 4 years of monocropping were significantly different from those in other years (Figure S1A,B). The difference in the rhizosphere fungal and bacterial microbiome between healthy and diseased American ginseng was greater after 4 years of monocropping (p < 0.05) compared to 1, 2, and 3 years of monocropping (Figure 4A,B).
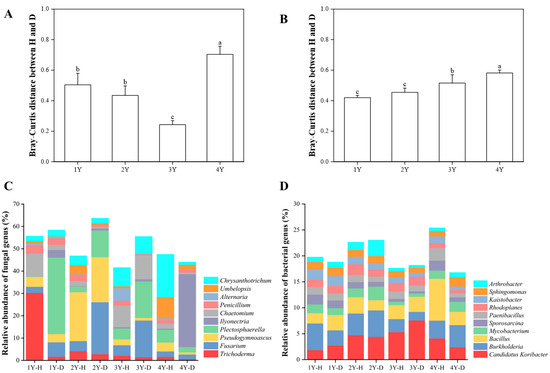
Figure 4.
The Bray–Curtis distance of fungal (A) and bacterial (B) microbiomes between healthy and diseased American ginseng when grown in soils with different monocropping years; and the relative abundance of the main fungal (C) and bacterial (D) groups present in the rhizosphere soil of the healthy and diseased American ginseng in 1–4 years of continuous monocropping (1Y, 2Y, 3Y, and 4Y) at the genus level; only the top ten genera in relative abundance are displayed. Different letters above the bars represent significant differences at p < 0.05 according to Duncan’s multiple range test.
The dominant fungal phylum of healthy and diseased rhizosphere soil belonged to Ascomycota (17.49–47.48%) (Table S1). Compared with the healthy American ginseng, the relative abundances of Ascomycota in the rhizosphere soil of diseased plants reduced after 1 and 4 years of monocropping, whereas they increased after 2 and 3 years.
A further annotation at the fungal genus level illustrated that the dominant genera were Fusarium spp. (2.10% to 25.88%), Trichoderma spp. (0.60% to 31.33%), and Ilyonectria spp. (0.03% to 32.65%). The relative abundance of Trichoderma spp. in the rhizosphere soil of the diseased plants was lower than that in the healthy American ginseng in 1–4 monocropping years. Compared with the diseased American ginseng, the relative abundance of Fusarium spp. of the healthy plant in 1, 2, and 3 years decreased by 3.74%, 18.79%, and 11.73%. In addition, after 4 years of monocropping, Ilyonectria spp. in the rhizosphere soil of the diseased plant was highly enriched compared with the healthy plant (0.42% vs. 32.65%).
The Proteobacteria (34.27% to 48.83%) and Actinobacteria (19.07% to 33.74%) were the dominant bacterial phyla in all the samples. The relative abundance of Actinobacteria in the healthy plants was higher than in diseased plants in 1, 2, and 3 years of monocropping (Table S2). The relative abundance of Actinobacteria gradually decreased with the increase in monocropping years. For bacterial genera, Candidatus Koribacter spp., Bacillus spp., and Burkholderia spp. were dominant in the rhizosphere soil. Compared with the healthy American ginseng, the relative abundance of Candidatus Koribacter spp. in rhizosphere soil of the diseased plants increased by 0.90% and 2.24% in 1 and 3 years of monocropping, respectively, but decreased by 0.34% and 1.69% in 2 and 4 years of monocropping, respectively. There was no significant difference between the Bacillus spp. and Burkholderia spp. between healthy and diseased American ginseng (Figure 4D).
2.4. Relationship between Phenolic Acids and Pathogens Abundance
The Pearson correlation was used to analyze the relationship between pathogen abundance and phenolic acids. The results illustrated that the abundance of F. solani had a significantly negative correlation with the content of sinapic acid (p = 0.026 < 0.05), p-coumaric acid (p = 0.022 < 0.05), and total phenolic acids (p = 0.025 < 0.05) (Figure 5). Additionally, the I. destructans abundance was negatively correlated with salicylic acid (p = 0.042 < 0.05) (Figure 5). However, the abundances of A. alternata and F. oxysporum were not associated with the content of phenolic acids.
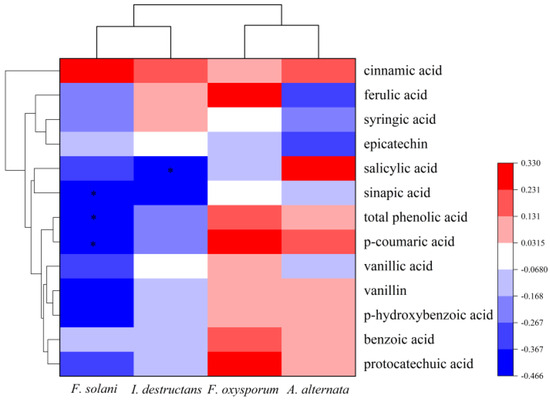
Figure 5.
The correlations between phenolic acids with pathogens abundance. * indicates a significant difference at p < 0.05.
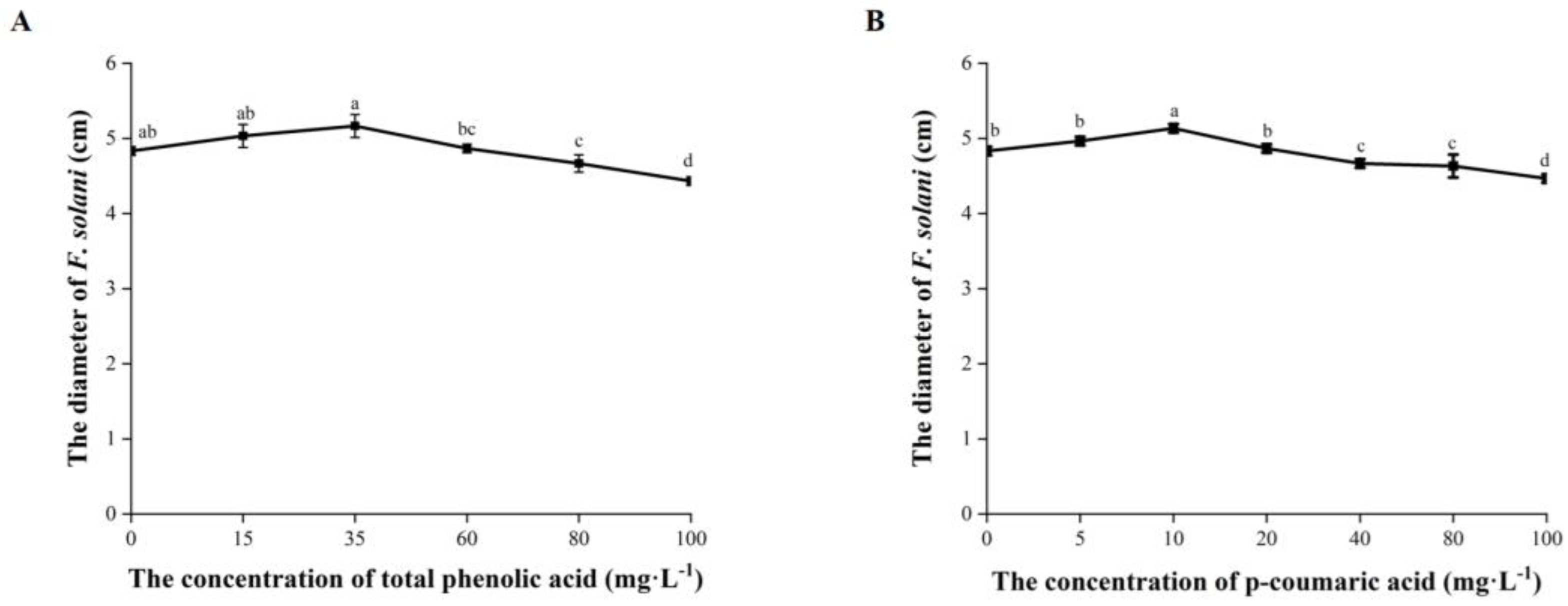
The mycelial growth of F. solani was significantly inhibited when the total phenolic acid was at a concentration ≥ 80 mg·L−1 with response indexes of 3.45% to 8.28% (Figure 6A). F. solani was significantly inhibited by p-coumaric acid at ≥40 mg·L−1 with response indexes of 3.45% to 7.59% (Figure 6B).
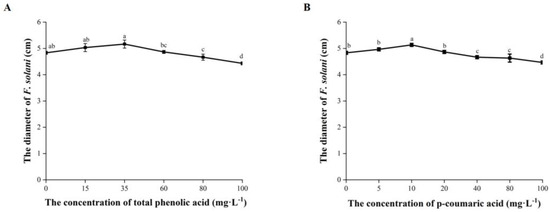
Figure 6.
Effects of total phenolic acid and p-coumaric acid on the mycelial growth of F. solani (A,B) at different concentrations. Error bars indicate the standard errors. Different letters above the bars within the same phenolic acid represent significant differences at p < 0.05 according to Duncan’s multiple range test.
3. Discussion
3.1. Potential Pathogens for Root Rot of American Ginseng on the Different Monocropping Years
Our study demonstrated that F. solani and I. destructans were the potential pathogens for American ginseng root rot, indicated by their higher abundances in the rhizosphere soil of diseased plants compared with healthy plants after 4-year monoculture. Previous findings showed that the pathogens that caused root rot in American ginseng mainly included Fusarium spp., Cylindrospora spp., and Ilyonectria spp. [25,26]. F. solani was a common plant pathogen that caused root rot and crown rot in potato [27], ginseng [28], and peanut [29]. Our results were consistent with the research in P. notoginseng rhizosphere soil, which found that the relative abundance of Ilyonectria spp. in 1- and 3-year-old plants was significantly higher than that in the 2-year-old plants, while the relative abundance of Fusarium spp. was highest in the rhizosphere soil of the 2-year-old plants [30]. Another study revealed that the relative abundance of Ilyonectria spp. in the rhizosphere soil of root rot of P. notoginseng was up to 75.90% to 80.10%, but it was not successfully isolated, possibly because of the slow growth characteristics [31].
3.2. The Changes in Phenolic Acid in the Healthy and Diseased American Ginseng during Various Consecutive Years of Monocropping
The phenolic acids in the soil were from the decomposition of plant residues and root exudation, which affected the growth and germination of American ginseng seedlings [32]. In our study, the difference in the total phenolic acid between the diseased and healthy American ginseng decreased gradually with the increase in monocropping years. The phenolic acid content in the healthy plants was larger than diseased plants after short-term monocropping, probably because the phenolic acid in the diseased American ginseng was degraded into resistant substances such as lignin [33]. Lignin can strengthen the cell walls and hinder pathogen invasion [34]. After long-term monocropping, there was no difference in phenolic acid content in the rhizosphere soil between healthy and diseased American ginseng, possibly because the root of the diseased plants decayed after a long-term monocropping and released a large quantity of phenolic acid into the soil to compensate the reduction consumed for resisting the pathogen.
The phenolic acids profiles in the rhizosphere soil of both healthy and diseased American ginseng were dominated by p-coumaric acid, which was consistent with the result in P. notoginseng [22]. P-coumaric acid is an important phenolic acid in the phenylalanine metabolism pathway of plants as the precursor of lignin synthesis and plays a significant role in pathogen resistance [35]. Islam et al. [36] found that p-coumaric acid promoted the growth of the plant cell wall and induced resistance in Chinese cabbage (Brassica rapa var. pekinensis) against the pathogens of Xanthomonas campestris pv. Campestris. Since p-coumaric acid in the rhizosphere soil had the highest percentage of the total, the trend of its change across the monocropping years was closely correlated with the total phenolic acid.
3.3. The Change in Microbial Community in the Healthy and Diseased American Ginseng during Various Consecutive Years of Monocropping
The composition of the bacterial and fungal microbiome between healthy and diseased American ginseng differed to a greater extent with the increase in monocropping year. Previous studies also observed the different structures of rhizosphere microbiome between the diseased and healthy plants in monocultured cotton [37], hybrid bamboo (Bambusa pervariabilis × Dendrocalamopsis daii) [38], and monkshood (Aconitum carmichaelii Debx.) in a 2-year monoculture field [39]. Unlike the above studies focusing on a single growing season, our finding confirmed the microbiome composition difference between healthy and diseased plants was related to the different monocropping years or monocropping history.
The difference in alpha diversity between healthy and diseased American ginseng gradually increased with the increase in monocropping years. With the increase in monocropping years, the pathogen’s abundance in the rhizosphere soil of diseased plants was higher in comparison to healthy plants. The pathogens could compete with other microorganisms for carbon sources and niches in rhizosphere soil, resulting in a decrease in microbiome diversity [40]. In addition, compared with the diseased American ginseng, more diverse root exudates secreted by the healthy plants provided a large range of carbon sources for microorganisms to enhance their diversity [41].
We found that the abundance of Fusarium spp. in the rhizosphere soil of diseased American ginseng was always higher than healthy plants in all the 1–4 years of monocropping history. However, the result of qPCR revealed that the abundance of F. solani in the rhizosphere soil of diseased American ginseng was significantly higher than the healthy plant after only 4 years of monocropping, and there was no difference in F. oxysporum between diseased and healthy plant. These results indicate that the Fusarium spp. genus included not only pathogens species but also nonpathogenic species. For example, a previous study showed that F. oxysporum strains isolated from the healthy plants of tomato (Solanum lycopersicum L.) were beneficial microbes for plant growth and resistance to pathogens [42]. In addition, we found that compared with the diseased American ginseng, the relative abundance of Trichoderma spp. in the rhizosphere soil of the healthy plant increased consistently across different monocropping histories. The Thichoderma spp. includes some species as biocontrol strains, which compete with pathogens in soil [43]; it uses enzymes to dissolve the pathogen’s cell wall, including cellulase, β-1,3-glucanase, and chitinase [44]. Meanwhile, the defense enzymes in plants, such as peroxidase (POD), phenylalanine ammonia lyase (PAL), and polyphenol oxidase (PPO), can be induced by Trichoderma spp. to resist the pathogen invasion and reduce the disease occurrence [45]. Trichoderma spp. also released ammonia, siderophores, auxin, and some secondary metabolites, which could promote the growth of plants [46,47].
3.4. The Phenolic Acid Suppressed the Growth of Pathogen
The increased occurrence of F. solani was associated with the decrease in total phenolic acid and p-coumaric acid. A similar finding was also reported previously that compared to the soil without a history of American ginseng cropping, the content of phenolic acid in the rhizosphere soil after 3 years of monocropping decreased, while pathogens, such as Helicoma spp., Cyberlinkera spp., and Saitozyma spp., increased [24]. Meanwhile, p-coumaric acid could also inhibit the germination of spores and sporulation of F. oxysporum [48]. Through bioassay, we found that the total phenolic acid at 80–100 mg·L−1 and p-coumaric acid at 40–100 mg·L−1 suppressed the growth rate of F. solani in vitro. The Fusarium wilt of cucumber (Cucumis sativus L.) was reduced by applying phenolic acid to soils, and with the increased concentration, the inhibition effect was enhanced [49]. Another study found that the mycelial growth and sporulation of F. oxysporum, the pathogen isolated from the rhizosphere soil of watermelon (Citrullus lanatus L.), was inhibited by cinnamic acid at concentrations between 0 and 1600 mg·L−1 [50].
4. Materials and Methods
4.1. Sample Collection
The long-term field experiment was conducted at Weihai city of Shandong Province, China (37°2′ N, 122°1′ E), using a split-plot design with American ginseng monocropping history (1–4 monocropping years) as the main plot and root disease infection as the subplot under 3 blocks (replicates). Samples were collected in different monocropping years. Plant roots were carefully dug out and shaken off to remove the soil that was not closely bound to the root. The soil that was tightly bound to the root was considered the rhizosphere soil. In each plot, 24 plants were sampled, and their rhizosphere soil was pooled as one sample. Finally, the 8 treatments included 1-year monocropping healthy rhizosphere soil (1Y-H), 1-year monocropping diseased rhizosphere soil (1Y-D), 2-year monocropping healthy rhizosphere soil (2Y-H), 2-year monocropping diseased rhizosphere soil (2Y-D), 3-year monocropping healthy rhizosphere soil (3Y-H), 3-year monocropping diseased rhizosphere soil (3Y-D), 4-year monocropping healthy rhizosphere soil (4Y-H), and 4-year monocropping diseased rhizosphere soil (4Y-D). Rhizosphere soils used for the determination of phenolic acid content were stored at 4 °C, and the 0.25 g of soil was stored at −80 °C to extract soil DNA.
4.2. The Extraction and Measurement of Phenolic Acid
According to the method of Meng et al. [51], the phenolic acids in the rhizosphere soil were extracted. Twenty grams of air-dried rhizosphere soil was placed in 500 mL conical flasks, and 150 mL of 2 mol·L−1 NaOH solution was added to each flask. The flasks were left for 24 h, then shaken at 210 r·min−1 for 30 min, centrifuged at 8000× g for 10 min, and the suspension was acidified to pH 2.5 with 6 mol·L−1 HCl, and the equal volume of ethyl acetate was added for extraction 3 times. The supernatant was mixed and concentrated by rotary steaming. The phenolic acids were redissolved in 1–2 mL 80% methanol, then filtered (0.22 μm). The filtrate was put into the injection bottle and stored at 4 °C for measurement.
Twelve compounds of phenolic acids, including protocatechuic acid, epicatechin, p-hydroxybenzoic acid, vanillic acid, p-coumaric acid, sinapic acid, vanillin, ferulic acid, benzoic acid, salicylic acid, syringic acid, and cinnamon acid, were analyzed by HPLC. The standards of the 12 phenolic acids (Sangon Biotech, Shanghai, China) were prepared at 200 mg·L−1.
The Agilent 1260 infinity II HPLC (Agilent Technologies Inc., Santa Clara, California, USA) was used to detect phenolic acid extracts. A Symmetry C-18 reversed column (4.6 mm × 250 mm, 5 µm) was installed in the HPLC, and the 5 µL sample was added into the column at 35 °C. The chromatograms of the sinapic acid, benzoic acid, salicylic acid, and ferulic acid were measured at 230 nm, and the chromatograms of the other phenolic acids were measured at 280 nm [52]. The 0.5% acetic water (A) and methanol (C) were used as the mobile phase, and the flow rate was set to 0.7 mL∙min−1 (A:C = 60:40, V:V). The separation of individual phenolic acids and total phenolic acids was performed by isometric elution and gradient elution, respectively. The following were the chromatographic conditions: First, 0–10 min, 95–80% A and 5–20% C; second, 10–13 min, 80–65% A and 20–35% C; then 13–33 min, 65–50% A and 35–50% C; and then 33–43 min, 50–5% A and 50–95% C; final, 43–53 min, 5–95% A and 95–5% C [53].
4.3. Soil DNA Extraction and Sequencing
The DNA was extracted by using DNeasy PowerSoil DNA Elution kit (Qiagen, Germantown, Maryland, USA). The extraction was carried out from 0.25 g rhizosphere soil samples based on the manufacturer’s instructions. The DNA integrity was detected by 1% agarose gel electrophoresis. The universal primers 341F/806R (341F: 5′-CCTAYGGGRBGCASCAG-3′/806R: 5′-GGACTACNNGGGTATCTAAT-3′) were used to amplify the V3-V4 region of the bacterial 16S rRNA gene, and the universal primers ITS1F/ITS2R (ITS1F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′/ITS2R: 5′-GCTGCGTTCTTCATCGATGC-3′) were used to amplify the fungal ITS region. The PCR product was sequenced by the Illumina Novaseq 6000 sequencing system (Illumina, Santiago, CA, USA). The rest of the extracted DNA was stored at −20 °C for quantitative PCR (qPCR) analysis.
The primer removal, quality filtering, denoising, splicing, chimera removal, and singleton removal were performed using DADA2 from QIIME2 (https://view.qiime2.org) (accessed on 22 May 2023) [54]. The amplicon sequence variants (ASVs) at the 99% similarity level were obtained by clustering the clean sequences [55]. Then the ASV sequences were aligned using the QIIME2 feature-classifier plugin to a pre-trained UNITE (release 8.0) [56] and Greengenes (release 13.8) [57] databases. Finally, at each level of classification—domain, phylum, class, order, family, genus, and species—the community composition of each sample was established. The QIIME2 feature-table plugin was used to filter any contaminating mitochondrial and chloroplast sequences.
4.4. The qPCR Analysis
The abundance of Fusarium solani, Fusarium oxysporum, Alternaria alternata, and Ilyonectria destructans was quantified by qPCR in a C1000 thermal cycler with a CFX96 real-time system (Bio-Rad Laboratories, Hercules, California, USA) to quantify the abundance of Fusarium solani, Fusarium oxysporum, Alternaria alternata, and Ilyonectria destructans. A total of 1 µL of DNA template, 10 µL of 2×SYBR Premix Ex Taq (Accurate Biology, Changsha, China), 8 µL of sterile distilled water, and 0.5 µmol·L−1 of each primer, including F. solani (ITS1F-F: 5′-CTTGGTCATTTAGAGGAGTAA-3′, AFP346-R: 5′-GGTATGTTCACAGGGTTGATG-3′) [58], F. oxysporum (ITS1F-F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′, AFP308-R: 5′-CGAATTAACGCGAGTCCCAAC-3′) [46], A. alternata (Alt-F: 5′-TGTCTTTTGCGTACTTCTTGTTTCCT-3′, Alt-R: 5′-CGACTTGTGCTGCGCTC-3′) [59], and I. destructans (Ily-F: 5′-GCTACCCTATAGCGCAGGTG-3′, Ily-R: 5′-CCGTACTGGCTGAAGAGTCA-3′) [59], were added to a total volume of 20 µl for the qPCR. The qPCR thermal conditions were 3 s at 95 °C, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, followed by melting curve analysis after the last cycle. The target fragment was cloned on a plasmid to prepare standards for qPCR. The standard curve was generated via 10 times gradient dilution of the plasmid DNA inserted with specific fragments according to the above protocol, and the efficiencies of the qPCR were 81.90–106.50%, and for all the standard curves, the coefficient of determination (R2) was >0.90.
4.5. Effects of Exogenous Phenolic Acid Concentration on Pathogen Suppression
Based on Durairaj’s pathogen list and methodology [60], American ginseng with root rot was collected from the fields in order to isolate pathogenic fungi. Pathogenicity tests were performed by inoculating the isolated strain on the puncture site of the surface of healthy American ginseng root. It was observed that the healthy American ginseng root exhibited symptoms similar to those on root rot plants observed in the field after inoculation. Finally, the isolates were identified as strains of Fusarium solani by morphological and molecular identification and were considered the target pathogens in this study.
According to the type and content of phenolic acids in the rhizosphere soil determined by HPLC in this study, total phenolic acids and p-coumaric acid were used to test their concentration on pathogen suppression. The solutions were added to PDA medium after being sterilized with a 0.22 μm filter membrane (Sangon, Shanghai, China). The total phenolic acid (1–2 years and 3–4 years) were at final concentrations of 15, 35, 60, 80, and 100 mg·L−1 (the 35 mg include 1.5 mg protocatechin, 3 mg p-hydroxybenzoic acid, 0.5 mg epicatechin, 2.5 mg vanillic acid, 3 mg syringic acid, 1.5 mg vanillin, 15 mg p-coumaric acid, 1.5 mg sinapic acid, 3 mg ferulic acid, 1 mg benzoic acid, 2.5 mg salicylic acid, and 0.2 mg cinnamic acid, and the other concentrations increase or decrease proportionally). The p-coumaric acid was at final concentrations of 5, 10, 20, 40, 80, and 100 mg·L−1. Mycelial discs (5 mm diameter each) of pure cultures of F. solani were incubated on PDA plates with various concentrations of phenolic acid at 25 °C for 6 days. Colony growth was evaluated by measuring the diameters based on the “cross” method [61].
4.6. Statistical Analysis
The core-diversity plugin within QIIME2 was used to calculate the Shannon index. Beta-diversity analysis of fungal and bacterial microbiomes in the rhizosphere soil was performed using the Bray–Curtis distance according to the “Vegan” package in R (version 4.2.1) [62]. Variable important in projection (VIP) from “mixOmics” in R (version 4.2.1) [63] and random forest (version 4.2.1) [64] was used to measure the importance of individual phenolic acids, determining the variation between experimental treatments. The relationship between the content of each phenolic acid and the abundance of pathogenic microorganisms was assessed by the Pearson correlation. SPSS 25.0 software (SPSS Inc., Chicago, IL, USA) was used to perform a single-factor analysis of variance (ANOVA) and Duncan multiple interval test (p < 0.05), and the homogeneity of variance was checked before statistical analysis.
5. Conclusions
Our results demonstrate that the pathogenic infection of American ginseng significantly influenced the secretion of phenolic acids and the assembly of the rhizosphere microbiome and that the monocropping history played a central role in modulating the interactions between disease infection, root exudation, and the diversity and structure of the microbial community in the rhizosphere soil.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12162993/s1, Figure S1: Principal component analysis of American ginseng rhizosphere fungi (A) and bacteria (B) in soils with healthy (H) and diseased (D) American ginseng in 1–4 years of continuous monocropping (1Y, 2Y, 3Y, and 4Y); Table S1: Relative abundance of fungi in the rhizosphere soil of diseased and healthy American ginseng at the phylum level; Table S2: Relative abundance of bacteria in the rhizosphere soil of diseased and healthy American ginseng at the phylum level.
Author Contributions
Conceptualization, Y.W. (Yanli Wei), H.L. and Y.Z.; methodology, J.L. and Y.W. (Yuanzheng Wu); software, J.H. and Y.Z.; formal analysis, Z.Z., Y.Z., and H.Y.; data curation, J.Z.; writing—original draft preparation, J.Z. and Y.W. (Yanli Wei); writing—review and editing, Y.Z.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Provincial Key Research and Development Project (major scientific and technological innovation) from Shandong Province (Project ID: 2020CXGC010803), International Cooperation Project of Science, Education, and Industry Integration Innovation Pilot Project from Shandong Academy of Sciences (project ID: 2022GH011, 2020KJC-GH07), Independent training innovation team project from Jinan city (project ID: 2020GXRC003).
Data Availability Statement
All sequencing raw data were uploaded to the Genome Sequence Ar-chive (GSA) database.
Acknowledgments
We thank the editor and the reviewers for their relevant comments that greatly improved the manuscript. We acknowledge all of our colleagues who worked on the field site and the funding support of our sponsors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ji, L.; Nasir, F.; Tian, L.; Chang, J.; Sun, Y.; Zhang, J.; Li, X.; Tian, C. Outbreaks of Root Rot Disease in Different Aged American Ginseng Plants Are Associated With Field Microbial Dynamics. Front. Microbiol. 2021, 12, 676880. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xu, J.; Zhang, L.; Yang, J.; Liao, B.; Li, X.; Chen, S. High-Throughput Sequencing Technology Reveals That Continuous Cropping of American Ginseng Results in Changes in the Microbial Community in Arable Soil. Chin. Med. 2017, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Singh, J.P. Long-Term Effects of Continuous Cropping and Different Nutrient Management Practices on the Distribution of Organic Nitrogen in Soil under Rice-Wheat System. Plant Soil Environ. 2014, 60, 63–68. [Google Scholar] [CrossRef]
- Van Wyk, D.A.B.; Adeleke, R.; Rhode, O.H.J.; Bezuidenhout, C.C.; Mienie, C. Ecological Guild and Enzyme Activities of Rhizosphere Soil Microbial Communities Associated with Bt-Maize Cultivation under Field Conditions in North West Province of South Africa. J. Basic Microbiol. 2017, 57, 781–792. [Google Scholar] [CrossRef]
- Neils, A.L.; Brisco-McCann, E.I.; Harlan, B.R.; Hausbeck, M.K. Management Strategies for Alternaria Leaf Blight on American Ginseng. Crop Prot. 2021, 139, 105302. [Google Scholar] [CrossRef]
- Bao, L.; Liu, Y.; Ding, Y.; Shang, J.; Wei, Y.; Tan, Y.; Zi, F. Interactions Between Phenolic Acids and Microorganisms in Rhizospheric Soil From Continuous Cropping of Panax notoginseng. Front. Microbiol. 2022, 13, 791603. [Google Scholar] [CrossRef]
- Goodwin, P.H. The Rhizosphere Microbiome of Ginseng. Microorganisms 2022, 10, 1152. [Google Scholar] [CrossRef]
- Steinauer, K.; Thakur, M.P.; Emilia Hannula, S.; Weinhold, A.; Uthe, H.; van Dam, N.M.; Martijn Bezemer, T. Root Exudates and Rhizosphere Microbiomes Jointly Determine Temporal Shifts in Plant-soil Feedbacks. Plant Cell Environ. 2023, 46, 1885–1899. [Google Scholar] [CrossRef]
- Guan, Y.M.; Ma, Y.Y.; Zhang, L.L.; Pan, X.X.; Liu, N.; Zhang, Y.Y. Occurrence of Sclerotinia Sclerotiorum Causing Sclerotinia Root Rot on American Ginseng in Northeastern China. Plant Dis. 2022, 106, 1518. [Google Scholar] [CrossRef]
- Schmidt, J.H.; Theisgen, L.V.; Finckh, M.R.; Šišić, A. Increased Resilience of Peas Toward Root Rot Pathogens Can Be Predicted by the Nematode Metabolic Footprint. Front. Sustain. Food Syst. 2022, 6, 881520. [Google Scholar] [CrossRef]
- Wu, Z.; Hao, Z.; Zeng, Y.; Guo, L.; Huang, L.; Chen, B. Molecular Characterization of Microbial Communities in the Rhizosphere Soils and Roots of Diseased and Healthy Panax Notoginseng. Antonie Van Leeuwenhoek 2015, 108, 1059–1074. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Riera, N.; Jin, T.; Li, J.; Wang, N. Huanglongbing Impairs the Rhizosphere-to-Rhizoplane Enrichment Process of the Citrus Root-Associated Microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef]
- Ahmad, T.; Farooq, S.; Mirza, D.N.; Kumar, A.; Mir, R.A.; Riyaz-Ul-Hassan, S. Insights into the Endophytic Bacterial Microbiome of Crocus Sativus: Functional Characterization Leads to Potential Agents That Enhance the Plant Growth, Productivity, and Key Metabolite Content. Microb. Ecol. 2022, 83, 669–688. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Cui, Y.; Li, H.; Kuang, A.; Li, X.; Wei, Y.; Ji, X. Rhizospheric Soil and Root Endogenous Fungal Diversity and Composition in Response to Continuous Panax Notoginseng Cropping Practices. Microbiol. Res. 2017, 194, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xu, J.; Feng, G.; Li, X.; Chen, S. Soil Bacterial and Fungal Community Dynamics in Relation to Panax Notoginseng Death Rate in a Continuous Cropping System. Sci. Rep. 2016, 6, 31802. [Google Scholar] [CrossRef]
- Tong, A.-Z.; Liu, W.; Liu, Q.; Xia, G.-Q.; Zhu, J.-Y. Diversity and Composition of the Panax Ginseng Rhizosphere Microbiome in Various Cultivation Modesand Ages. BMC Microbiol. 2021, 21, 18. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, Q.; Liu, X.; Liu, L.; Ding, W. Plant Growth Promoting Rhizobacteria Alleviate Aluminum Toxicity and Ginger Bacterial Wilt in Acidic Continuous Cropping Soil. Front. Microbiol. 2020, 11, 569512. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial Community Composition Is Related to Soil Biological and Chemical Properties and Bacterial Wilt Outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, J.; Pan, D.; Ge, X.; Jin, X.; Chen, S.; Wu, F. P-Coumaric Can Alter the Composition of Cucumber Rhizosphere Microbial Communities and Induce Negative Plant-Microbial Interactions. Biol. Fertil. Soils 2018, 54, 363–372. [Google Scholar] [CrossRef]
- Su, L.; Zhang, L.; Nie, D.; Kuramae, E.E.; Shen, B.; Shen, Q. Bacterial Tomato Pathogen Ralstonia Solanacearum Invasion Modulates Rhizosphere Compounds and Facilitates the Cascade Effect of Fungal Pathogen Fusarium Solani. Microorganisms 2020, 8, 806. [Google Scholar] [CrossRef]
- Zhao, Y.-M.; Cheng, Y.-X.; Ma, Y.-N.; Chen, C.-J.; Xu, F.-R.; Dong, X. Role of Phenolic Acids from the Rhizosphere Soils of Panax Notoginseng as a Double-Edge Sword in the Occurrence of Root-Rot Disease. Molecules 2018, 23, 819. [Google Scholar] [CrossRef] [PubMed]
- He, C.N.; Gao, W.W.; Yang, J.X.; Bi, W.; Zhang, X.S.; Zhao, Y.J. Identification of Autotoxic Compounds from Fibrous Roots of Panax quinquefolium L. Plant Soil 2009, 318, 63–72. [Google Scholar] [CrossRef]
- An, S.; Wei, Y.; Li, H.; Zhao, Z.; Hu, J.; Philp, J.; Ryder, M.; Toh, R.; Li, J.; Zhou, Y.; et al. Long-Term Monocultures of American Ginseng Change the Rhizosphere Microbiome by Reducing Phenolic Acids in Soil. Agriculture 2022, 12, 640. [Google Scholar] [CrossRef]
- DesRochers, N.; Walsh, J.P.; Renaud, J.B.; Seifert, K.A.; Yeung, K.K.-C.; Sumarah, M.W. Metabolomic Profiling of Fungal Pathogens Responsible for Root Rot in American Ginseng. Metabolites 2020, 10, 35. [Google Scholar] [CrossRef]
- Zhou, X.; Luo, C.; Li, K.; Zhu, D.; Jiang, L.; Wu, L.; Li, Y.; He, X.; Du, Y. First Report of Fusarium Striatum Causing Root Rot Disease of Panax Notoginseng in Yunnan, China. Phyton 2022, 91, 13–20. [Google Scholar] [CrossRef]
- Chehri, K.; Ghasempour, H.R.; Karimi, N. Molecular Phylogenetic and Pathogenetic Characterization of Fusarium Solani Species Complex (FSSC), the Cause of Dry Rot on Potato in Iran. Microb. Pathog. 2014, 67, 14–19. [Google Scholar] [CrossRef]
- Li, T.; Kim, J.-H.; Jung, B.; Ji, S.; Seo, M.W.; Han, Y.K.; Lee, S.W.; Bae, Y.S.; Choi, H.-G.; Lee, S.-H.; et al. Transcriptome Analyses of the Ginseng Root Rot Pathogens Cylindrocarpon Destructans and Fusarium Solani to Identify Radicicol Resistance Mechanisms. J. Ginseng Res. 2020, 44, 161–167. [Google Scholar] [CrossRef]
- Erazo, J.G.; Palacios, S.A.; Pastor, N.; Giordano, F.D.; Rovera, M.; Reynoso, M.M.; Venisse, J.S.; Torres, A.M. Biocontrol Mechanisms of Trichoderma Harzianum ITEM 3636 against Peanut Brown Root Rot Caused by Fusarium Solani RC 386. Biol. Control 2021, 164, 104774. [Google Scholar] [CrossRef]
- Wang, B.; Xia, Q.; Li, Y.; Zhao, J.; Yang, S.; Wei, F.; Huang, X.; Zhang, J.; Cai, Z. Root Rot-Infected Sanqi Ginseng Rhizosphere Harbors Dynamically Pathogenic Microbiotas Driven by the Shift of Phenolic Acids. Plant Soil 2021, 465, 385–402. [Google Scholar] [CrossRef]
- Mi, C.; Yang, R.; Rao, J.; Yang, S.; Wei, F.; Li, O.; Hu, X. Unveiling of Dominant Fungal Pathogens Associated With Rusty Root Rot of Panax Notoginseng Based on Multiple Methods. Plant Dis. 2017, 101, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, L.; Dong, K.; Dong, Y.; Xiao, J. Cinnamic Acid Inhibited Growth of Faba Bean and Promoted the Incidence of Fusarium Wilt. Plants 2018, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Li, D.; Zhou, C.; Wu, Y.; Miao, P.; Dong, Q.; Zhu, S.; Pan, C. Application of Insecticides on Peppermint (Mentha × Piperita L.) Induces Lignin Accumulation in Leaves by Consuming Phenolic Acids and Thus Potentially Deteriorates Quality. J. Plant Physiol. 2022, 279, 153836. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic Acids Act as Signaling Molecules in Plant-Microbe Symbioses. Plant Signal. Behav. 2010, 5, 359–368. [Google Scholar] [CrossRef]
- Hellinger, J.; Kim, H.; Ralph, J.; Karlen, S.D. P -Coumaroylation of Lignin Occurs Outside of Commelinid Monocots in the Eudicot Genus Morus (Mulberry). Plant Physiol. 2023, 191, 854–861. [Google Scholar] [CrossRef]
- Islam, M.T.; Lee, B.-R.; Das, P.R.; La, V.H.; Jung, H.; Kim, T.-H. Characterization of P-Coumaric Acid-Induced Soluble and Cell Wall-Bound Phenolic Metabolites in Relation to Disease Resistance to Xanthomonas Campestris Pv. Campestris in Chinese Cabbage. Plant Physiol. Biochem. 2018, 125, 172–177. [Google Scholar] [CrossRef]
- Wei, F.; Feng, H.; Zhang, D.; Feng, Z.; Zhao, L.; Zhang, Y.; Deakin, G.; Peng, J.; Zhu, H.; Xu, X. Composition of Rhizosphere Microbial Communities Associated With Healthy and Verticillium Wilt Diseased Cotton Plants. Front. Microbiol. 2021, 12, 618169. [Google Scholar] [CrossRef]
- Lin, T.; Li, L.; Gu, X.; Owusu, A.M.; Li, S.; Han, S.; Cao, G.; Zhu, T.; Li, S. Seasonal Variations in the Composition and Diversity of Rhizosphere Soil Microbiome of Bamboo Plants as Infected by Soil-Borne Pathogen and Screening of Associated Antagonistic Strains. Ind. Crops Prod. 2023, 197, 116641. [Google Scholar] [CrossRef]
- Li, Y.; He, F.; Guo, Q.; Feng, Z.; Zhang, M.; Ji, C.; Xue, Q.; Lai, H. Compositional and Functional Comparison on the Rhizosphere Microbial Community between Healthy and Sclerotium Rolfsii-Infected Monkshood (Aconitum Carmichaelii) Revealed the Biocontrol Potential of Healthy Monkshood Rhizosphere Microorganisms. Biol. Control 2022, 165, 104790. [Google Scholar] [CrossRef]
- Zhang, M.; Jia, J.; Lu, H.; Feng, M.; Yang, W. Functional Diversity of Soil Microbial Communities in Response to Supplementing 50% of the Mineral N Fertilizer with Organic Fertilizer in an Oat Field. J. Integr. Agric. 2021, 20, 2255–2264. [Google Scholar] [CrossRef]
- Wutzler, T.; Reichstein, M. Priming and Substrate Quality Interactions in Soil Organic Matter Models. Biogeosciences 2013, 10, 2089–2103. [Google Scholar] [CrossRef]
- Olivain, C.; Humbert, C.; Nahalkova, J.; Fatehi, J.; L’Haridon, F.; Alabouvette, C. Colonization of Tomato Root by Pathogenic and Nonpathogenic Fusarium Oxysporum Strains Inoculated Together and Separately into the Soil. Appl. Environ. Microbiol. 2006, 72, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Han, M.; Hu, Y.; Li, Z.; Liu, C.; Wang, X.; Tian, Q.; Jiao, W.; Hu, J.; Liu, L.; et al. Effects of Continuous Cropping of Sweet Potato on the Fungal Community Structure in Rhizospheric Soil. Front. Microbiol. 2019, 10, 2269. [Google Scholar] [CrossRef] [PubMed]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma Species—Opportunistic, Avirulent Plant Symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- López, A.C.; Giorgio, E.M.; Vereschuk, M.L.; Zapata, P.D.; Luna, M.F.; Alvarenga, A.E. Ilex Paraguariensis Hosts Root-Trichoderma Spp. with Plant-Growth-Promoting Traits: Characterization as Biological Control Agents and Biofertilizers. Curr. Microbiol. 2023, 80, 120. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, W.; Mao, Y.; Ma, S.; Chen, X.; Shen, X.; Yin, C.; Mao, Z. Isolation of Trichoderma Virens 6PS-2 and Its Effects on Fusarium Proliferatum f. Sp. Malus Domestica MR5 Related to Apple Replant Disease (ARD) in China. Hortic. Plant J. 2022, in press, S246801412200111X. [Google Scholar] [CrossRef]
- Ren, L.; Huo, H.; Zhang, F.; Hao, W.; Xiao, L.; Dong, C.; Xu, G. The Components of Rice and Watermelon Root Exudates and Their Effects on Pathogenic Fungus and Watermelon Defense. Plant Signal. Behav. 2016, 11, e1187357. [Google Scholar] [CrossRef]
- Yuan, F.; Zhang, C. lan Alleviating Effect of Phenol Compounds on Cucumber Fusarium Wilt and Mechanism. Agric. Sci. China 2003, 6, 60–65. [Google Scholar]
- Wu, H.-S.; Raza, W.; Fan, J.-Q.; Sun, Y.-G.; Bao, W.; Shen, Q.-R. Cinnamic Acid Inhibits Growth but Stimulates Production of Pathogenesis Factors by in Vitro Cultures of Fusarium Oxysporum f.Sp. Niveum. J. Agric. Food Chem. 2008, 56, 1316–1321. [Google Scholar] [CrossRef]
- Meng, L.; Xia, Z.; Lv, J.; Liu, G.; Tan, Y.; Li, Q. Extraction and GC-MS Analysis of Phenolic Acids in Rhizosphere Soil of Pinellia Ternate. J. Radiat. Res. Appl. Sci. 2022, 15, 40–45. [Google Scholar] [CrossRef]
- Rahman, M.; Punja, Z.K. Biochemistry of Ginseng Root Tissues Affected by Rusty Root Symptoms. Plant Physiol. Biochem. 2005, 43, 1103–1114. [Google Scholar] [CrossRef]
- Das, S.; Sultana, K.W.; Chandra, I. Adventitious Rhizogenesis in Basilicum Polystachyon (L.) Moench Callus and HPLC Analysis of Phenolic Acids. Acta Physiol. Plant 2021, 43, 146. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ Preprints 2018, e27295v1. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Larsson, K.-H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A Database Providing Web-Based Methods for the Molecular Identification of Ectomycorrhizal Fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Lievens, B.; Brouwer, M.; Vanachter, A.C.R.C.; Levesque, C.A.; Cammue, B.P.A.; Thomma, B.P.H.J. Quantitative Assessment of Phytopathogenic Fungi in Various Substrates Using a DNA Macroarray. Environ. Microbiol. 2005, 7, 1698–1710. [Google Scholar] [CrossRef]
- Pavón, M.Á.; González, I.; Rojas, M.; Pegels, N.; Martín, R.; García, T. PCR Detection of Alternaria Spp. in Processed Foods, Based on the Internal Transcribed Spacer Genetic Marker. J. Food Prot. 2011, 74, 240–247. [Google Scholar] [CrossRef]
- Durairaj, K.; Velmurugan, P.; Park, J.-H.; Chang, W.-S.; Park, Y.-J.; Senthilkumar, P.; Choi, K.-M.; Lee, J.-H.; Oh, B.-T. An Investigation of Biocontrol Activity Pseudomonas and Bacillus Strains against Panax Ginseng Root Rot Fungal Phytopathogens. Biol. Control. 2018, 125, 138–146. [Google Scholar] [CrossRef]
- Luo, Y.; Tian, P. Growth and Characteristics of Two Different Epichloë Sinensis Strains Under Different Cultures. Front. Microbiol. 2021, 12, 726935. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A Tool for Visualizing High-Throughput Microbial Community Data. GigaSci 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Wang, Z.; Wang, S.; Zhang, J.; Yu, J. Locating Facial Landmarks Using Probabilistic Random Forest. IEEE Signal Process. Lett. 2015, 22, 2324–2328. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).