Cytokinin Promotes Jasmonic Acid Accumulation in the Control of Maize Leaf Growth
Abstract
1. Introduction
2. Results
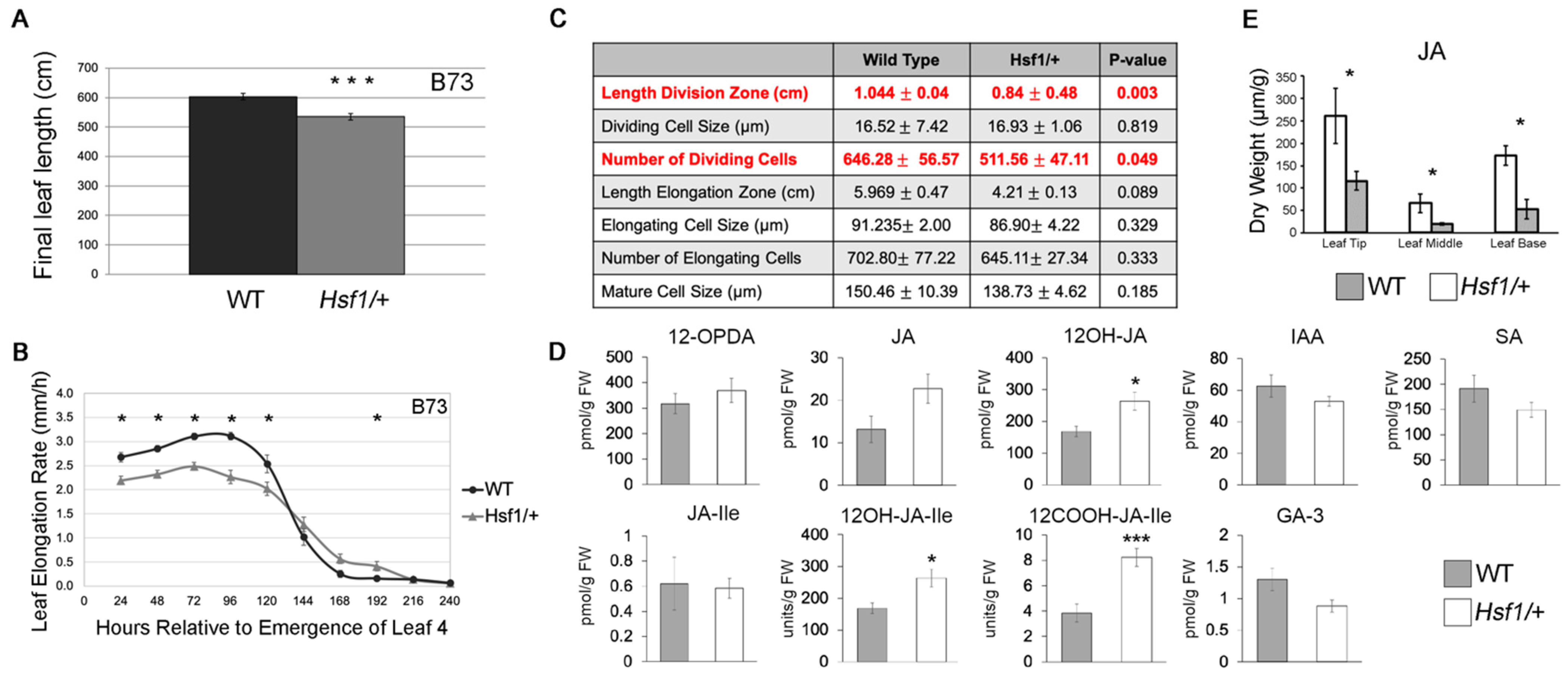
2.1. Hsf1 Mutants Have a Reduced Leaf Growth Phenotype
2.2. Distinct Jasmonates Accumulate in Growing Hsf1/+ Maize Leaves
2.3. JA Pathway Genes Are Upregulated in the Leaf Growth Zone of Hsf1 Mutants
2.4. Exogenous Jasmonic Acid Treatments Reduce Leaf Growth Rate in Maize
2.5. Hsf1 Is Less Responsive to Exogenous Jasmonic Acid Treatment
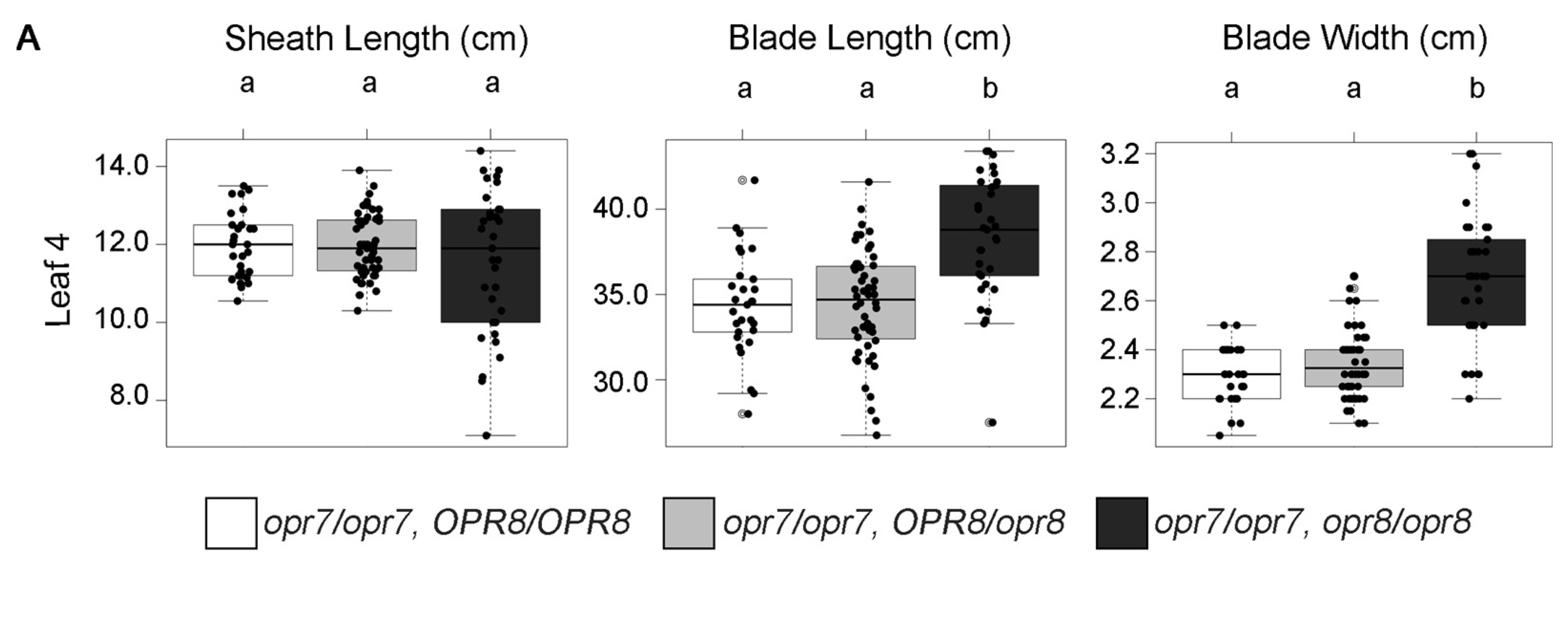
2.6. Growth Is Enhanced in Jasmonic Acid-Deficient Mutants
2.7. JA-Deficient Mutants Suppress the Reduced Leaf Growth Phenotype in Hsf1 Mutants
2.8. Exogenous CK Treatment Induces Expression of JA Pathway Genes in the Leaf Growth Zone
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Material, Genetics, Phenotypic Measurements, and Analysis
5.2. Standard Germinating Seed Hormone Treatment
5.3. Cytokinin
5.4. Jasmonic Acid
5.5. Final Leaf Size Measurements and Kinematic Analysis
5.6. Growth Rate Measurement
5.7. Seedling Treatments and JA-Pathway Gene Expression Analysis
5.8. Plant Metabolite Assays
5.9. Statistics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conklin, P.A.; Strable, J.; Li, S.; Scanlon, M.J. On the Mechanisms of Development in Monocot and Eudicot Leaves. New Phytol. 2019, 221, 706–724. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.D.; Nath, U. On the Evolution of Developmental Mechanisms: Divergent Polarities in Leaf Growth as a Case Study. Plant Signal Behav. 2016, 11, e1126030. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nelissen, H.; Gonzalez, N.; Inzé, D. Leaf Growth in Dicots and Monocots: So Different yet so Alike. Curr. Opin. Plant Biol. 2016, 33, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Frébort, I.; Kowalska, M.; Hluska, T.; Frébortová, J.; Galuszka, P. Evolution of Cytokinin Biosynthesis and Degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent Advances and Emerging Trends in Plant Hormone Signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Wolters, H.; Jürgens, G. Survival of the Flexible: Hormonal Growth Control and Adaptation in Plant Development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef]
- Liu, H.; Timko, M.P. Jasmonic Acid Signaling and Molecular Crosstalk with Other Phytohormones. Int. J. Mol. Sci. 2021, 22, 2914. [Google Scholar] [CrossRef]
- Hou, X.; Ding, L.; Yu, H. Crosstalk between GA and JA Signaling Mediates Plant Growth and Defense. Plant Cell Rep. 2013, 32, 1067–1074. [Google Scholar] [CrossRef]
- Huang, Y.; Han, C.; Peng, W.; Peng, Z.; Xiong, X.; Zhu, Q.; Gao, B.; Xie, D.; Ren, C. Brassinosteroid Negatively Regulates Jasmonate Inhibition of Root Growth in Arabidopsis [Review of Brassinosteroid Negatively Regulates Jasmonate Inhibition of Root Growth in Arabidopsis. Plant Signal Behav. 2010, 5, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Han, C.; Peng, W.; Huang, Y.; Peng, Z.; Xiong, X.; Zhu, Q.; Gao, B.; Xie, D. A Leaky Mutation in DWARF4 Reveals an Antagonistic Role of Brassinosteroid in the Inhibition of Root Growth by Jasmonate in Arabidopsis. Plant Physiol. 2009, 151, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A Genomic Approach to Identify Regulatory Nodes in the Transcriptional Network of Systemic Acquired Resistance in Plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic Acid Inhibits Pathogen Growth in Plants through Repression of the Auxin Signaling Pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef]
- Miller, C.O.; Skoog, F.; Saltza, M.H.; Strong, F.M. Kinetin, A Cell Division Factor From Deoxyribonucleic Acid1. J. Am. Chem. Soc. 1955, 77, 1392. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of Plant Growth by Cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [PubMed]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis Cytokinin Receptor Mutants Reveal Functions in Shoot Growth, Leaf Senescence, Seed Size, Germination, Root Development, and Cytokinin Metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Jensen, H.; Novák, O.; Strnad, M.; Werner, T.; Schmülling, T. Gain-of-Function Mutants of the Cytokinin Receptors AHK2 and AHK3 Regulate Plant Organ Size, Flowering Time and Plant Longevity. Plant Physiol. 2017, 173, 1783–1797. [Google Scholar] [CrossRef]
- Burr, C.A.; Sun, J.; Yamburenko, M.V.; Willoughby, A.; Hodgens, C.; Boeshore, S.L.; Elmore, A.; Atkinson, J.; Nimchuk, Z.L.; Bishopp, A.; et al. The HK5 and HK6 Cytokinin Receptors Mediate Diverse Developmental Pathways in Rice. Development 2020, 147, dev191734. [Google Scholar] [CrossRef]
- Lomin, S.N.; Yonekura-Sakakibara, K.; Romanov, G.A.; Sakakibara, H. Ligand-Binding Properties and Subcellular Localization of Maize Cytokinin Receptors. J. Exp. Bot. 2011, 62, 5149–5159. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural Basis for Cytokinin Receptor Signaling: An Evolutionary Approach. Plant Cell Rep. 2013, 32, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Bertrand-Garcia, R.; Freeling, M. Hairy-Sheath Frayed #1-o: A Systemic, Heterochronic Mutant of Maize That Specifies Slow Developmental Stage Transitions. Am. J. Bot. 1991, 78, 747–765. [Google Scholar]
- Muszynski, M.G.; Moss-Taylor, L.; Chudalayandi, S.; Cahill, J.; Del Valle-Echevarria, A.R.; Alvarez-Castro, I.; Petefish, A.; Sakakibara, H.; Krivosheev, D.M.; Lomin, S.N.; et al. The Maize Hairy Sheath Frayed1 (Hsf1) Mutation Alters Leaf Patterning through Increased Cytokinin Signaling. Plant Cell 2020, 32, 1501–1518. [Google Scholar] [CrossRef] [PubMed]
- Pineda Rodo, A.; Brugière, N.; Vankova, R.; Malbeck, J.; Olson, J.M.; Haines, S.C.; Martin, R.C.; Habben, J.E.; Mok, D.W.S.; Mok, M.C. Over-Expression of a Zeatin O-Glucosylation Gene in Maize Leads to Growth Retardation and Tasselseed Formation. J. Exp. Bot. 2008, 59, 2673–2686. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Huang, P.-C.; Borrego, E.; Kolomiets, M. New Perspectives into Jasmonate Roles in Maize. Plant Signal Behav. 2014, 9, e970442. [Google Scholar] [CrossRef]
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate Biosynthesis and Signaling in Monocots: A Comparative Overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 Catalyze Two Successive Oxidation Steps of Plant Hormone Jasmonoyl-Isoleucine for Catabolic Turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef]
- Kitaoka, N.; Matsubara, T.; Sato, M.; Takahashi, K.; Wakuta, S.; Kawaide, H.; Matsui, H.; Nabeta, K.; Matsuura, H. Arabidopsis CYP94B3 Encodes Jasmonyl-L-Isoleucine 12-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Jasmonate. Plant Cell Physiol. 2011, 52, 1757–1765. [Google Scholar] [CrossRef]
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic Reticulum-Associated Inactivation of the Hormone Jasmonoyl-L-Isoleucine by Multiple Members of the Cytochrome P450 94 Family in Arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738. [Google Scholar] [CrossRef]
- Koo, A.J.K.; Howe, G.A. Catabolism and Deactivation of the Lipid-Derived Hormone Jasmonoyl-Isoleucine. Front. Plant Sci. 2012, 3, 19. [Google Scholar] [CrossRef]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, Signaling, and Transport of Jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Lunde, C.; Kimberlin, A.; Leiboff, S.; Koo, A.J.; Hake, S. Tasselseed5 Overexpresses a Wound-Inducible Enzyme, ZmCYP94B1, That Affects Jasmonate Catabolism, Sex Determination, and Plant Architecture in Maize. Commun. Biol. 2019, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.K.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 Mediates Catabolism and Inactivation of the Plant Hormone Jasmonoyl-L-Isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.N.; Holtsclaw, R.E.; Kimberlin, A.; Sen, S.; Zeng, S.; Joshi, T.; Lei, Z.; Sumner, L.W.; Singh, K.; Matsuura, H.; et al. 12-Hydroxy-Jasmonoyl-l-Isoleucine Is an Active Jasmonate That Signals through CORONATINE INSENSITIVE 1 and Contributes to the Wound Response in Arabidopsis. Plant Cell Physiol. 2019, 60, 2152–2166. [Google Scholar] [CrossRef] [PubMed]
- Acosta, I.F.; Laparra, H.; Romero, S.P.; Schmelz, E.; Hamberg, M.; Mottinger, J.P.; Moreno, M.A.; Dellaporta, S.L. Tasselseed1 Is a Lipoxygenase Affecting Jasmonic Acid Signaling in Sex Determination of Maize. Science 2009, 323, 262–265. [Google Scholar] [CrossRef]
- Yan, Y.; Christensen, S.; Isakeit, T.; Engelberth, J.; Meeley, R.; Hayward, A.; Emery, R.J.N.; Kolomiets, M.V. Disruption of OPR7 and OPR8 Reveals the Versatile Functions of Jasmonic Acid in Maize Development and Defense. Plant Cell 2012, 24, 1420–1436. [Google Scholar] [CrossRef]
- Yamane, H.; Sugawara, J.; Suzuki, Y.; Shimamura, E.; Takahashi, N. Syntheses of Jasmonic Acid Related Compounds and Their Structure-Activity Relationships on the Growth of Rice Seedlings. Agric. Biol. Chem. 1980, 44, 2857–2864. [Google Scholar]
- Noir, S.; Bömer, M.; Takahashi, N.; Ishida, T.; Tsui, T.-L.; Balbi, V.; Shanahan, H.; Sugimoto, K.; Devoto, A. Jasmonate Controls Leaf Growth by Repressing Cell Proliferation and the Onset of Endoreduplication While Maintaining a Potential Stand-by Mode. Plant Physiol. 2013, 161, 1930–1951. [Google Scholar] [CrossRef]
- Zhang, Y.; Turner, J.G. Wound-Induced Endogenous Jasmonates Stunt Plant Growth by Inhibiting Mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef]
- Nelissen, H.; Rymen, B.; Coppens, F.; Dhondt, S.; Fiorani, F.; Beemster, G.T.S. Kinematic Analysis of Cell Division in Leaves of Mono- and Dicotyledonous Species: A Basis for Understanding Growth and Developing Refined Molecular Sampling Strategies. In Plant Organogenesis: Methods and Protocols; Smet, I., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 247–264. [Google Scholar]
- Nelissen, H.; Rymen, B.; Jikumaru, Y.; Demuynck, K.; Van Lijsebettens, M.; Kamiya, Y.; Inzé, D.; Beemster, G.T.S. A Local Maximum in Gibberellin Levels Regulates Maize Leaf Growth by Spatial Control of Cell Division. Curr. Biol. 2012, 22, 1183–1187. [Google Scholar] [CrossRef]
- Gao, X.; Starr, J.; Göbel, C.; Engelberth, J.; Feussner, I.; Tumlinson, J.; Kolomiets, M. Maize 9-Lipoxygenase ZmLOX3 Controls Development, Root-Specific Expression of Defense Genes, and Resistance to Root-Knot Nematodes. Mol. Plant Microbe Interact. 2008, 21, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.J.; Kieber, J.J. Cytokinin Signaling. Curr. Opin. Plant Biol. 2005, 8, 518–525. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.C.; Haberer, G.; Ferreira, F.J.; Deruère, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis Response Regulators Are Partially Redundant Negative Regulators of Cytokinin Signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic Expression of KNOTTED1-like Homeobox Protein Induces Expression of Cytokinin Biosynthesis Genes in Rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef]
- Dermastia, M.; Ravnikar, M.; Vilhar, B.; Kovac, M. Increased Level of Cytokinin Ribosides in Jasmonic Acid-Treated Potato (Solanum Tuberosum) Stem Node Cultures. Physiol. Plant 1994, 92, 241–246. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Benková, E. Cytokinin Cross-Talking during Biotic and Abiotic Stress Responses. Front. Plant Sci. 2013, 4, 451. [Google Scholar] [CrossRef]
- Ueda, J.; Kato, J. Inhibition of Cytokinin-Induced Plant Growth by Jasmonic Acid and Its Methyl Ester. Physiol. Plant 1982, 54, 249–252. [Google Scholar] [CrossRef]
- Sun, X.; Cahill, J.; Van Hautegem, T.; Feys, K.; Whipple, C.; Novák, O.; Delbare, S.; Versteele, C.; Demuynck, K.; De Block, J.; et al. Altered Expression of Maize PLASTOCHRON1 Enhances Biomass and Seed Yield by Extending Cell Division Duration. Nat. Commun. 2017, 8, 14752. [Google Scholar] [CrossRef]
- Giulini, A.; Wang, J.; Jackson, D. Control of Phyllotaxy by the Cytokinin-Inducible Response Regulator Homologue ABPHYL1. Nature 2004, 430, 1031–1034. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schäfer, M.; Brütting, C.; Baldwin, I.T.; Kallenbach, M. High-Throughput Quantification of More than 100 Primary- and Secondary-Metabolites, and Phytohormones by a Single Solid-Phase Extraction Based Sample Preparation with Analysis by UHPLC-HESI-MS/MS. Plant Methods 2016, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2023. [Google Scholar]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uyehara, A.N.; Del Valle-Echevarria, A.R.; Hunter, C.T.; Nelissen, H.; Demuynck, K.; Cahill, J.F.; Gorman, Z.; Jander, G.; Muszynski, M.G. Cytokinin Promotes Jasmonic Acid Accumulation in the Control of Maize Leaf Growth. Plants 2023, 12, 3014. https://doi.org/10.3390/plants12163014
Uyehara AN, Del Valle-Echevarria AR, Hunter CT, Nelissen H, Demuynck K, Cahill JF, Gorman Z, Jander G, Muszynski MG. Cytokinin Promotes Jasmonic Acid Accumulation in the Control of Maize Leaf Growth. Plants. 2023; 12(16):3014. https://doi.org/10.3390/plants12163014
Chicago/Turabian StyleUyehara, Aimee N., Angel R. Del Valle-Echevarria, Charles T. Hunter, Hilde Nelissen, Kirin Demuynck, James F. Cahill, Zachary Gorman, Georg Jander, and Michael G. Muszynski. 2023. "Cytokinin Promotes Jasmonic Acid Accumulation in the Control of Maize Leaf Growth" Plants 12, no. 16: 3014. https://doi.org/10.3390/plants12163014
APA StyleUyehara, A. N., Del Valle-Echevarria, A. R., Hunter, C. T., Nelissen, H., Demuynck, K., Cahill, J. F., Gorman, Z., Jander, G., & Muszynski, M. G. (2023). Cytokinin Promotes Jasmonic Acid Accumulation in the Control of Maize Leaf Growth. Plants, 12(16), 3014. https://doi.org/10.3390/plants12163014





