Genome-Wide Analysis and Expression Profiling of Trehalose-6-Phosphate Phosphatase (TPP) in Punica granatum in Response to Abscisic-Acid-Mediated Drought Stress
Abstract
:1. Introduction
2. Results
2.1. Basic Information of TPP Family Genes in Pomegranate
2.2. Phylogenetic Tree of Full-Length Pomegranate, Arabidopsis, and Tomato TPP Proteins
2.3. Gene Structure and Motif Analysis
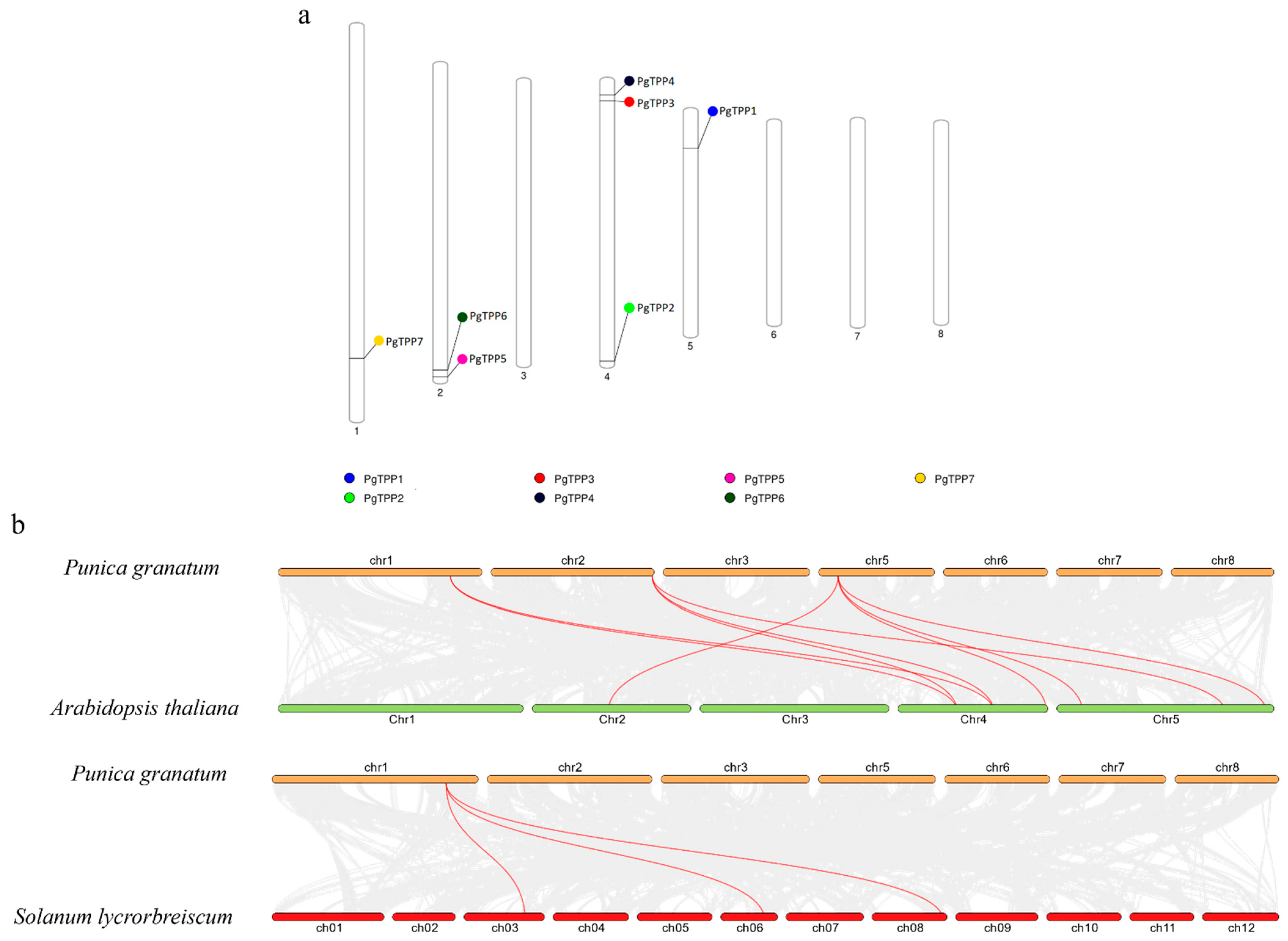
2.4. Chromosomal Distribution and Homology Analysis of PgTPP Genes
2.5. Cis-Acting Regulatory Elements in PgTPP Promoters
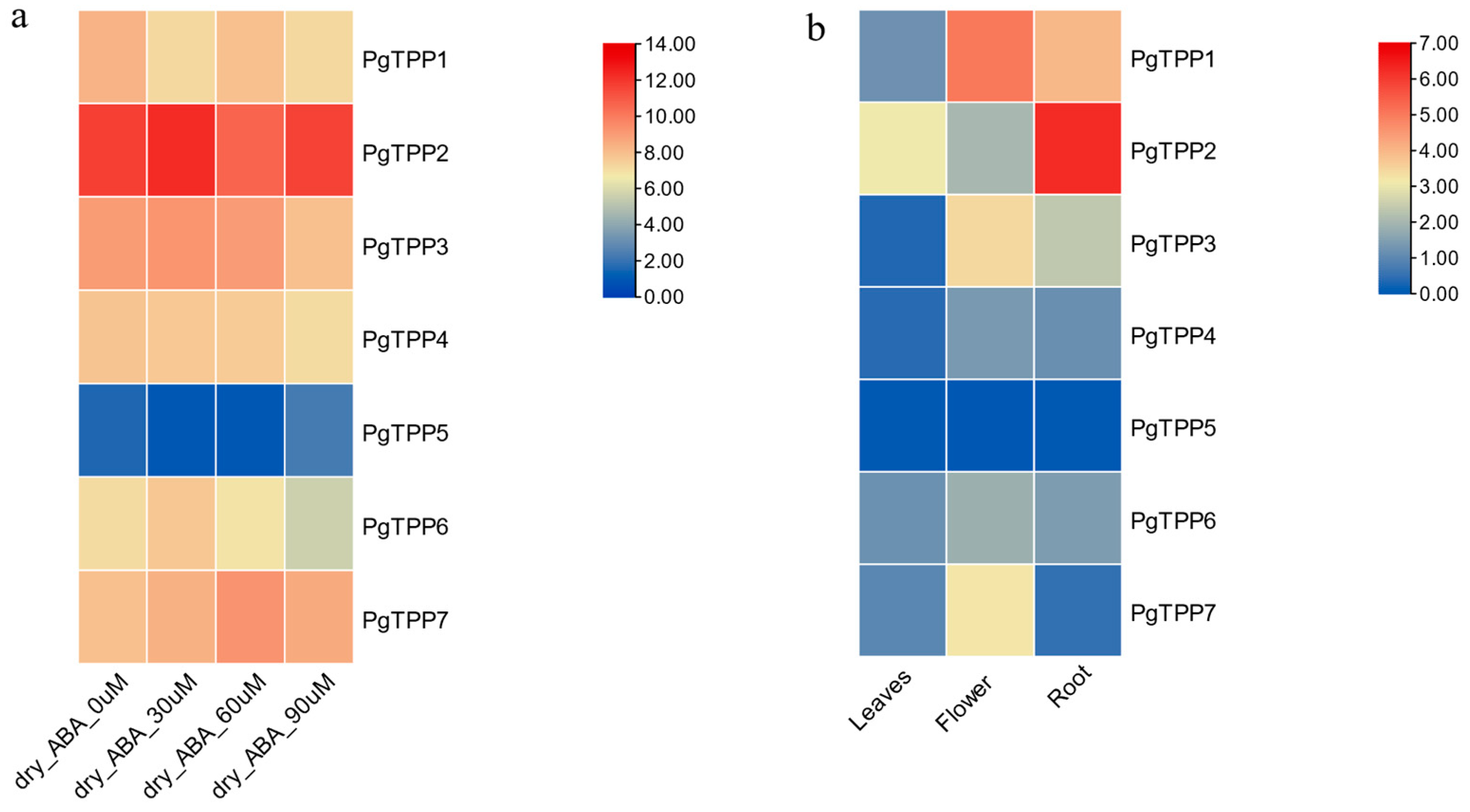
2.6. Expression Profiles of PgTPP Genes under Abscisic-Acid-Mediated Drought Stress
2.7. Tissue-Differential Gene Expression Patterns of PgTPPs
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification and Annotation of TPP Genes in Pomegranate
4.2. Phylogenetic Tree of Full-Length Pomegranate, Arabidopsis, and Tomato
4.3. Gene Structure and Conserved Motif Analysis
4.4. Chromosomal Distribution and Homology Analysis of TPP Genes
4.5. Prediction and Analysis of Cis-Acting Elements in Promoter Regions
4.6. Expression Profile Analysis of the TPP Gene Family in Different Tissues and in Leaves of Pomegranate under Abscisic-Acid-Mediated Drought Stress
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Paul, M.J.; Gonzalez-Uriarte, A.; Griffiths, C.A.; Hassani-Pak, K. The role of trehalose 6-phosphate in crop yield and resilience. Plant Physiol. 2018, 177, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Pandey, A.; Prasad, M.; Böhmer, M. Abiotic stress signaling in plants: Functional genomic intervention. Front. Plant Sci. 2016, 7, 681. [Google Scholar] [CrossRef]
- Fichtner, F.; Lunn, J.E. The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; López-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis. Plant Physiol. 2012, 160, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Wuyts, J.; Verschooten, K.; Vandesteene, L.; Van Dijck, P. The Cytophaga hutchinsonii ChTPSP: First characterized bifunctional TPS–TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins. Mol. Biol. Evol. 2010, 27, 359–369. [Google Scholar] [CrossRef]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef]
- Lin, Q.; Gong, J.; Zhang, Z.; Meng, Z.; Wang, J.; Wang, S.; Sun, J.; Gu, X.; Jin, Y.; Wu, T. The Arabidopsis thaliana trehalose-6-phosphate phosphatase gene AtTPPI regulates primary root growth and lateral root elongation. Front. Plant Sci. 2023, 13, 1088278. [Google Scholar] [CrossRef]
- Lin, Q.; Yang, J.; Wang, Q.; Zhu, H.; Chen, Z.; Dao, Y.; Wang, K. Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef]
- Shima, S.; Matsui, H.; Tahara, S.; Imai, R. Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J. 2007, 274, 1192–1201. [Google Scholar] [CrossRef]
- Pramanik, M.H.R.; Imai, R. Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol. Biol. 2005, 58, 751–762. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Pelayo, M.A.F.; Trijatmiko, K.R.; Gabunada, L.F.M.; Alam, R.; Jimenez, R.; Mendioro, M.S.; Slamet-Loedin, I.H.; Sreenivasulu, N.; Bailey-Serres, J. A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 2015, 1, 15124. [Google Scholar] [CrossRef]
- Du, L.; Li, S.; Ding, L.; Cheng, X.; Kang, Z.; Mao, H. Genome-wide analysis of trehalose-6-phosphate phosphatases (TPP) gene family in wheat indicates their roles in plant development and stress response. BMC Plant Biol. 2022, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Mollavali, M.; Börnke, F. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase genes of tomato (Solanum lycopersicum L.) and analysis of their differential expression in response to temperature. Int. J. Mol. Sci. 2022, 23, 11436. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, M.; Huang, Y.; Zhu, P.; Qian, G.; Zhang, Y.; Liu, Y.; Zhou, J.; Li, L. Genome-Wide Identification and Analysis of Stress Response of Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase Genes in Quinoa. Int. J. Mol. Sci. 2023, 24, 6950. [Google Scholar] [CrossRef]
- Fahmy, H.; Hegazi, N.; El-Shamy, S.; Farag, M.A. Pomegranate juice as a functional food: A comprehensive review of its polyphenols, therapeutic merits, and recent patents. Food Funct. 2020, 11, 5768–5781. [Google Scholar] [CrossRef] [PubMed]
- Bhantana, P.; Lazarovitch, N. Evapotranspiration, crop coefficient and growth of two young pomegranate (Punica granatum L.) varieties under salt stress. Agric. Water Manag. 2010, 97, 715–722. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Delorge, I.; Figueroa, C.M.; Feil, R.; Lunn, J.E.; Van Dijck, P. Trehalose-6-phosphate synthase 1 is not the only active TPS in Arabidopsis thaliana. Biochem. J. 2015, 466, 283–290. [Google Scholar] [CrossRef]
- Acosta-Pérez, P.; Camacho-Zamora, B.D.; Espinoza-Sánchez, E.A.; Gutiérrez-Soto, G.; Zavala-García, F.; Abraham-Juárez, M.J.; Sinagawa-García, S.R. Characterization of Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase genes and analysis of its differential expression in maize (Zea mays) seedlings under drought stress. Plants 2020, 9, 315. [Google Scholar] [CrossRef]
- Jin, Q.; Hu, X.; Li, X.; Wang, B.; Wang, Y.; Jiang, H.; Mattson, N.; Xu, Y. Genome-wide identification and evolution analysis of trehalose-6-phosphate synthase gene family in Nelumbo nucifera. Front. Plant Sci. 2016, 7, 1445. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A computational framework for the prediction of single-and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S. FastQC. A quality control tool for high throughput sequence data 2010, 370.Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]



| Locus_ID | Gene Name | Protein _ID | Transcript_ID | Location, Start–End | Strand | Gene Length | CDS (bp) | Protein Length (A.A) | Protein Molecular Weight (Da) | PI | GRAVY | No of Exons | Cellular Localization Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOC116206686 | PgTPP1 | XP_031395344.1 | XM_031539484.1 | Chromosome 5, NC_045131.1 (5264138..5267309) | − | 3172 | 1119 | 372 | 41,950 | 9.46 | −0.405 | 11 | Plastid |
| LOC116206238 | PgTPP2 | XP_031394912.1 | XM_031539052.1 | Chromosome 4 NC_045130.1 (39635185..39639127) | − | 3943 | 1149 | 382 | 42,707 | 9.04 | −0.283 | 12 | Plastid |
| LOC116206238 | PgTPP2 | XP_031394913.1 | XM_031539053.1 | Chromosome 4 NC_045130.1 (39635185..39639127) | − | 3943 | 1149 | 382 | 42,707 | 9.04 | −0.283 | 12 | Plastid |
| LOC116202804 | PgTPP3 | XP_031390275.1 | XM_031534415.1 | Chromosome 4 NC_045130.1 (2818814..2822149) | + | 3336 | 1092 | 363 | 40,638 | 9.3 | −0.291 | 11 | Nuclei |
| LOC116202804 | PgTPP3 | XP_031390276.1 | XM_031534416.1 | Chromosome 4 NC_045130.1 (2818814..2822149) | + | 3336 | 1083 | 360 | 40,283 | 9.24 | −0.304 | 11 | Nuclei |
| LOC116202804 | PgTPP3 | XP_031390277.1 | XM_031534417.1 | Chromosome 4 NC_045130.1 (2818814..2822149) | + | 3336 | 1035 | 344 | 38,591 | 9.44 | −0.313 | 10 | Nuclei |
| LOC116202714 | PgTPP4 | XP_031390179.1 | XM_031534319.1 | Chromosome 4 NC_045130.1 (2007596..2011818) | + | 4223 | 1149 | 382 | 42,190 | 6.17 | −0.318 | 14 | Mitochondria |
| LOC116202714 | PgTPP4 | XP_031390180.1 | XM_031534322.1 | Chromosome 4 NC_045130.1 (2007596..2011818) | + | 4223 | 1149 | 382 | 42,850 | 6.03 | −0.335 | 14 | Mitochondria |
| LOC116202714 | PgTPP4 | XP_031390181.1 | XM_031534320.1 | Chromosome 4 NC_045130.1 (2007596..2011818) | + | 4223 | 1149 | 382 | 42,850 | 6.03 | −0.335 | 14 | Mitochondria |
| LOC116202714 | PgTPP4 | XP_031390182.1 | XM_031534321.1 | Chromosome 4 NC_045130.1 (2007596..2011818) | + | 4223 | 1149 | 382 | 42,850 | 6.03 | −0.335 | 13 | Mitochondria |
| LOC116202714 | PgTPP4 | XP_031390184.1 | XM_031534324.1 | Chromosome 4 NC_045130.1 (2007596..2011818) | + | 4223 | 1131 | 376 | 42,850 | 6.03 | −0.335 | 14 | Mitochondria |
| LOC116194837 | PgTPP5 | XP_031379593.1 | XM_031523735.1 | Chromosome 2 NC_045128.1 (44074459..44078382) | − | 3924 | 948 | 315 | 35,674 | 9.31 | −0.37 | 13 | Plastid |
| LOC116194837 | PgTPP5 | XP_031379594.1 | XM_031523734.1 | Chromosome 2 NC_045128.1 (44074459..44078382 complement) | − | 3924 | 948 | 315 | 35,674 | 9.31 | −0.37 | 12 | Plastid |
| LOC116194837 | PgTPP5 | XP_031379595.1 | XM_031523733.1 | Chromosome 2 NC_045128.1 (44074459..44078382) | − | 3924 | 948 | 315 | 35,674 | 9.31 | −0.37 | 12 | Plastid |
| LOC116194202 | PgTPP6 | XP_031378813.1 | XM_031522953.1 | Chromosome 2 NC_045128.1 (43096590..43104600) | + | 8011 | 1113 | 370 | 41,565 | 9.16 | −0.4 | 10 | Golgi |
| LOC116193147 | PgTPP7 | XP_031377785.1 | XM_031521925.1 | Chromosome 1 NC_045127.1 (46957219..46962168) | + | 4950 | 1071 | 356 | 39,602 | 6.8 | −0.303 | 14 | Nuclei |
| LOC116193147 | PgTPP7 | XP_031377784.1 | XM_031521924.1 | Chromosome 1 NC_045127.1 (46957219..46962168) | + | 4950 | 1170 | 389 | 43,295 | 8.58 | −0.358 | 13 | Nuclei |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omari Alzahrani, F. Genome-Wide Analysis and Expression Profiling of Trehalose-6-Phosphate Phosphatase (TPP) in Punica granatum in Response to Abscisic-Acid-Mediated Drought Stress. Plants 2023, 12, 3076. https://doi.org/10.3390/plants12173076
Omari Alzahrani F. Genome-Wide Analysis and Expression Profiling of Trehalose-6-Phosphate Phosphatase (TPP) in Punica granatum in Response to Abscisic-Acid-Mediated Drought Stress. Plants. 2023; 12(17):3076. https://doi.org/10.3390/plants12173076
Chicago/Turabian StyleOmari Alzahrani, Fatima. 2023. "Genome-Wide Analysis and Expression Profiling of Trehalose-6-Phosphate Phosphatase (TPP) in Punica granatum in Response to Abscisic-Acid-Mediated Drought Stress" Plants 12, no. 17: 3076. https://doi.org/10.3390/plants12173076
APA StyleOmari Alzahrani, F. (2023). Genome-Wide Analysis and Expression Profiling of Trehalose-6-Phosphate Phosphatase (TPP) in Punica granatum in Response to Abscisic-Acid-Mediated Drought Stress. Plants, 12(17), 3076. https://doi.org/10.3390/plants12173076






