Genome-Wide Identification and Characterization of the PP2C Family from Zea mays and Its Role in Long-Distance Signaling
Abstract
:1. Introduction
2. Results
2.1. Identification of PP2C Members in the Maize Genome
2.2. Evolutionary and Comparative Analysis of the PP2C Family in Four Species
2.3. Gene Structure, Conserved Motif, and Evolutionary Analysis of ZmPP2Cs
2.4. Chromosomal Localization and Gene Duplication of ZmPP2C Gene Family
2.5. Cis-Element Analysis in the Promoters of ZmPP2Cs
2.6. Protein–Protein Interaction Network of ZmPP2Cs
2.7. Expression Profiles of ZmPP2C Genes in Different Maize Tissues
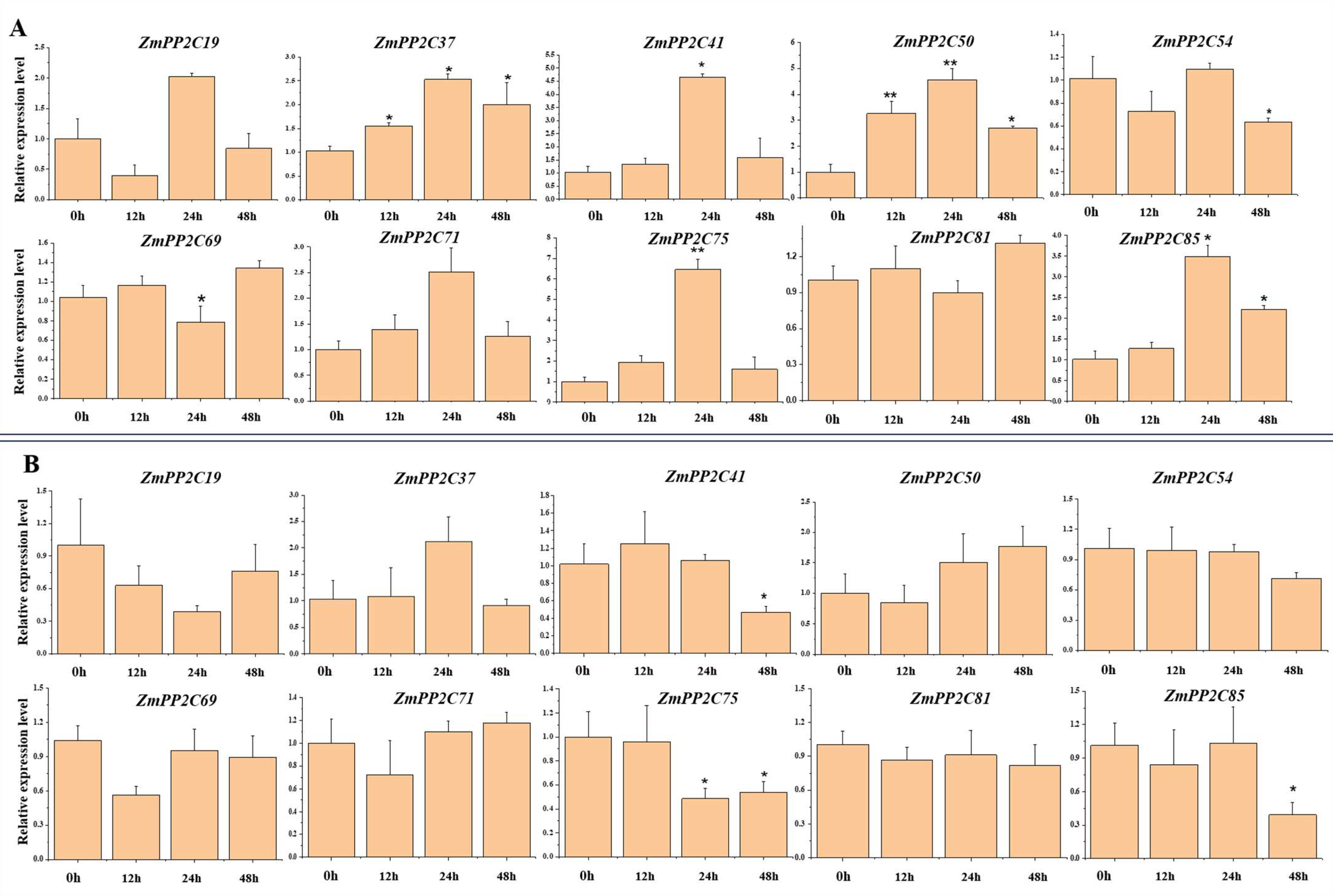
2.8. Expression of ZmPP2C Genes under Different Nitrogen Concentration Stress
2.9. Differential Gene Expression of the ZmPP2Cs under Heterogeneous Nitrogen Stress
2.10. Subcellular Localization
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Database Searches and Genome-Wide Identification of PP2C Family Members in Maize
5.2. Analysis of Maize PP2C Protein Feature
5.3. Evolutionary Tree, Gene Structure, and Conserved Motifs
5.4. Chromosomal Location and Synteny Correlation Analysis
5.5. Identification of Cis-Regulatory Elements in Promoters of ZmPP2Cs
5.6. Protein–Protein Network Interaction Analysis of ZmPP2C Genes
5.7. Expression Analysis of ZmPP2C Genes between Different Tissues
5.8. Plant Growth and Treatments
5.9. Split-Root Experiments
5.10. RNA Extraction and qRT-PCR
5.11. Subcellular Localization Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, H.; Ning, L.; He, B.; Wang, Y.; Ge, M.; Xu, J.; Zhao, H. Cross-species root transcriptional network analysis highlights conserved modules in response to nitrate between maize and sorghum. Int. J. Mol. Sci. 2020, 21, 1445. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutierrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Vidal, E.A.; Gutiérrez, R.A. Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 2012, 15, 185–191. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M.; Forde, B.G. Molecular and developmental biology of inorganic nitrogen nutrition. Arab. Book/Am. Soc. Plant Biol. 2002, 1, e0011. [Google Scholar] [CrossRef]
- Yu, P.; Li, X.; Yuan, L.; Li, C. A novel morphological response of maize (Zea mays) adult roots to heterogeneous nitrate supply revealed by a split-root experiment. Physiol. Plant. 2014, 150, 133–144. [Google Scholar] [CrossRef]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef]
- Tilman, D.; Clark, M. Food, agriculture & the environment: Can we feed the world & save the earth? Daedalus 2015, 144, 8–23. [Google Scholar]
- Wang, X.T.; Wang, W.; Chen, J.J.; Chu, C.C. Long-distance signal transduction of nitrogen and phosphorus in plants. J. S. China Agric. Univ. 2022, 43, 78–86. [Google Scholar]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Gansel, X.; Munos, S.; Tillard, P.; Gojon, A. Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: Relation with long-distance and local controls by N status of the plant. Plant J. 2001, 26, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Krouk, G.; Ristova, D.; Shasha, D.; Birnbaum, K.D.; Coruzzi, G.M. Nitrogen economics of root foraging: Transitive closure of the nitrate—Cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 2011, 108, 18524–18529. [Google Scholar] [CrossRef]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plant. 2017, 3, 1–6. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Ota, R.; Ohkubo, Y.; Yamashita, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 2020, 11, 641. [Google Scholar] [CrossRef]
- Ohkubo, Y.; Kuwata, K.; Matsubayashi, Y. A type 2C protein phosphatase activates high-affinity nitrate uptake by dephosphorylating NRT2. 1. Nat. Plants 2021, 7, 310–316. [Google Scholar] [CrossRef]
- Van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell. 2014, 26, 2367–2389. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Wong, M.M.; Verslues, P.E. The flip side of phospho-signalling: Regulation of protein dephosphorylation and the protein phosphatase 2Cs. Plant Cell Environ. 2019, 42, 2913–2930. [Google Scholar] [CrossRef]
- Cao, J.; Jiang, M.; Li, P.; Chu, Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016, 17, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989, 58, 453–508. [Google Scholar] [CrossRef] [PubMed]
- Kerk, D.; Templeton, G.; Moorhead, G.B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2018, 146, 351. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanism. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Moorhead, G.B.; Trinkle-Mulcahy, L.; Ulke-Lemée, A. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Bio. 2007, 8, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 1–18. [Google Scholar] [CrossRef]
- Sugimoto, H.; Kondo, S.; Tanaka, T.; Imamura, C.; Muramoto, N.; Hattori, E.; Ohto, C. Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J. EXP. BOT. 2014, 65, 5385–5400. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Wu, J.; Sun, X.; Dai, M. The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 2019, 20, 3573. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Gao, S.; Qin, F.; Dai, M. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol Plant. 2017, 10, 456–469. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, Q.; Shi, J.; Jin, X.; He, Y.; Zhang, Y.; He, G. Brachypodium distachyon BdPP2CA6 interacts with BdPYLs and BdSnRK2 and positively regulates salt tolerance in transgenic Arabidopsis. Front Plant Sci. 2017, 8, 264. [Google Scholar] [CrossRef]
- Vega, A.; Fredes, I.; O’Brien, J.; Shen, Z.; Ötvös, K.; Abualia, R.; Gutiérrez, R.A. Nitrate triggered phosphoproteome changes and a PIN2 phosphosite modulating root system architecture. EMBO Rep. 2021, 22, e51813. [Google Scholar] [CrossRef] [PubMed]
- Sathee, L.; Krishna, G.K.; Adavi, S.B.; Jha, S.K.; Jain, V. Role of protein phosphatases in the regulation of nitrogen nutrition in plants. Physiol. Mol. Bio. Plant. 2021, 27, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.; Ludewig, U.; Neuhäuser, B. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell. 2017, 29, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Léran, S.; Edel, K.H.; Pervent, M.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Lacombe, B. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 2015, 8, ra43. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Feng, S.; Amjad, H.; Letuma, P.; Zhan, W.; Li, Z.; Lin, W. Protein phosphatase (PP2C9) induces protein expression differentially to mediate nitrogen utilization efficiency in rice under nitrogen-deficient condition. Int. J. Mol. Sci. 2018, 19, 2827. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Pan, S. Maize protein phosphatase gene family: Identification and molecular characterization. BMC Genom. 2014, 15, 1–20. [Google Scholar] [CrossRef]
- Fan, K.; Yuan, S.; Chen, J.; Chen, Y.; Li, Z.; Lin, W.; Lin, W. Molecular evolution and lineage-specific expansion of the PP2C family in Zea mays. Planta 2019, 250, 1521–1538. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; von Mering, C. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic. Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 1–21. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Zhang, J.; Sun, J.; Zhang, Y.; Lu, M.; Hu, J. Genome-wide characterization of protein phosphatase 2C genes in Populus euphratica and their expression profiling under multiple abiotic stresses. Tree Genet. Genomes. 2018, 14, 1–13. [Google Scholar] [CrossRef]
- Bhalothia, P.; Lata, S.; Khan, Z.H.; Kumar, B.; Mehrotra, S.; Mehrotra, R. Genome wide analysis of protein phosphatase 2C (PP2C) genes in Glycine max and Sorghum bicolor. Curr. Biol. 2018, 7, 302–308. [Google Scholar] [CrossRef]
- Khan, N.; Ke, H.; Hu, C.M.; Naseri, E.; Haider, M.S.; Ayaz, A.; Hou, X. Genome-wide identification, evolution, and transcriptional profiling of PP2C gene family in Brassica rapa. Biomed. Res. Int. 2019, 2019, 2965035. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Xu, Z.; Wu, L.; Chen, X.; Cui, Y.; Wang, Y.; Yao, H. Genome-wide identification of protein phosphatase 2C family members in Glycyrrhiza uralensis Fisch. and their response to abscisic acid and polyethylene glycol stress. J. Taibah. Univ. Sci. 2021, 15, 1260–1268. [Google Scholar] [CrossRef]
- Cheng, C.; Cai, Z.; Su, R.; Zhong, Y.; Chen, L.; Wang, L.; Li, C. Genome-wide identification and gene expression analysis of Clade A protein phosphatase 2C family genes in Brassica juncea var tumida. BioRxiv 2021. [Google Scholar] [CrossRef]
- Hapgood, J.P.; Riedemann, J.; Scherer, S.D. Regulation of gene expression by GC-rich DNA Cis-elements. Cell Biol. Int. 2001, 25, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Knudsen, S. The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J. 1993, 4, 343–355. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, H.; Long, Y.; Shu, Y.; Zhai, J. PPRD: A comprehensive online database for expression analysis of~ 45,000 plant public RNA-Seq libraries. BioRxiv. 2022, 20, 806–808. [Google Scholar]
- Chen, L.; Wang, G.; Chen, P.; Zhu, H.; Wang, S.; Ding, Y. Shoot-root communication plays a key role in physiological alterations of rice (Oryza sativa) under iron deficiency. Front. Plant Sci. 2018, 9, 757. [Google Scholar] [CrossRef]
- Chen, L.; Lin, Y.; Chen, P.; Wang, S.; Ding, Y. Effect of Iron Deficiency on the Protein Profile of Rice (Oryza sativa) Phloem Sap. Chin. Bull. Bot. 2019, 54, 194. [Google Scholar]
- Xia, C.; Zheng, Y.; Huang, J.; Zhou, X.; Li, R.; Zha, M.; Zhang, C. Elucidation of the mechanisms of long-distance mRNA movement in a Nicotiana benthamiana/tomato heterograft system. Plant Physiol. 2018, 177, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Huang, J.; Lan, H.; Zhang, C. Long-distance movement of mineral deficiency-responsive mRNAs in Nicotiana benthamiana/tomato heterografts. Plants 2020, 9, 876. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sun, X.; Guo, Z.; Joldersma, D.; Guo, L.; Qiao, X.; Zhang, S. Genome-wide identification and evolution of the PP2C gene family in eight Rosaceae species and expression analysis under stress in Pyrus bretschneideri. Front. Genet. 2021, 12, 770014. [Google Scholar] [CrossRef]
- Haider, M.S.; Khan, N.; Pervaiz, T.; Zhongjie, L.; Nasim, M.; Jogaiah, S.; Jinggui, F. Genome-wide identification, evolution, and molecular characterization of the PP2C gene family in woodland strawberry. Gene 2019, 702, 27–35. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
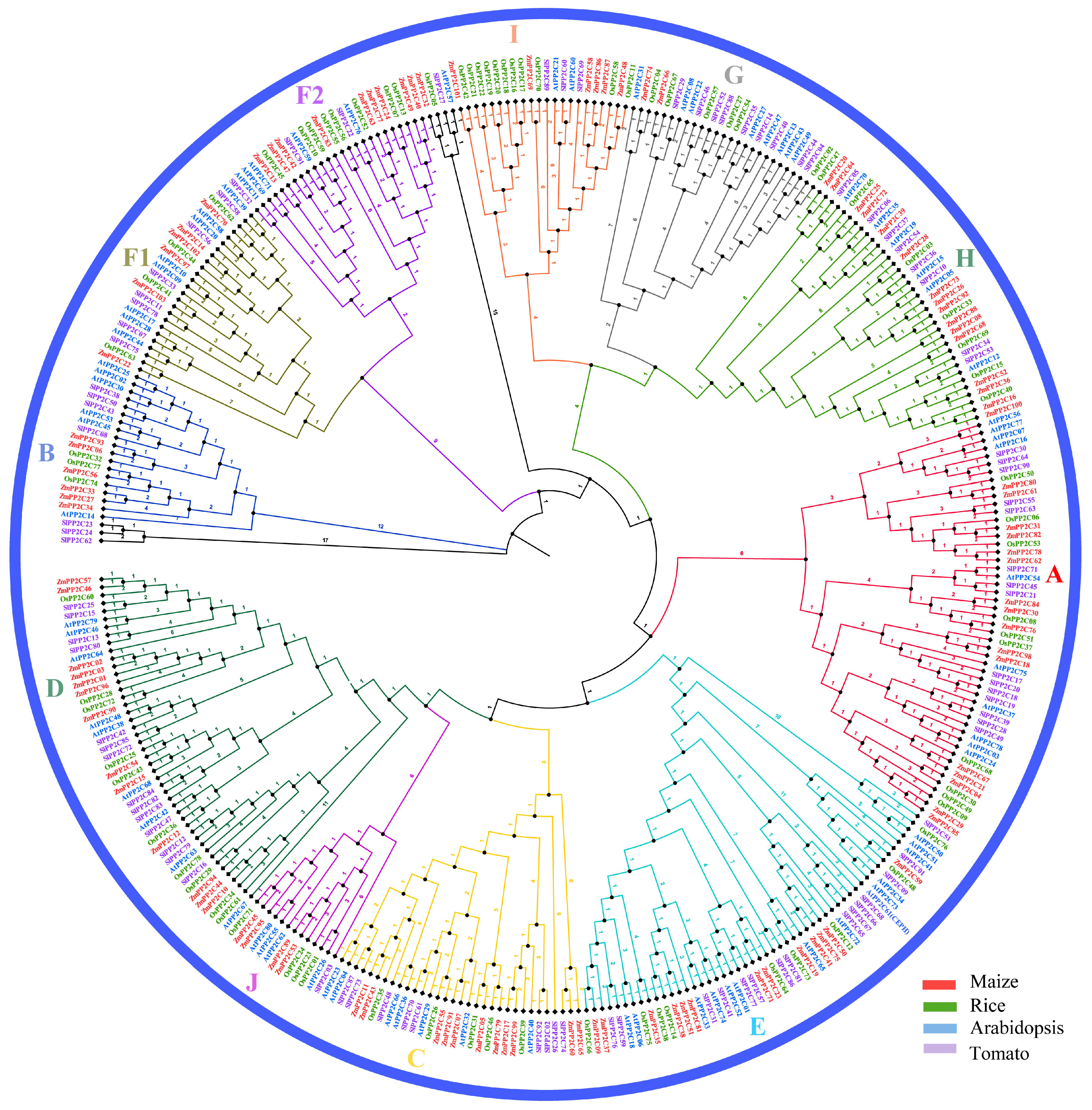







| Subgroup of PP2C Genes | Numbers of AtPP2Cs | Numbers of OsPP2Cs | Numbers of ZmPP2Cs | Numbers of SlPP2Cs |
|---|---|---|---|---|
| A | 10 | 10 | 16 | 15 |
| B | 6 | 3 | 6 | 4 |
| C | 7 | 5 | 11 | 8 |
| D | 9 | 10 | 13 | 14 |
| E | 14 | 9 | 13 | 15 |
| F1 | 8 | 4 | 6 | 7 |
| F2 | 5 | 8 | 9 | 3 |
| G | 8 | 7 | 2 | 9 |
| H | 6 | 6 | 16 | 8 |
| I | 2 | 11 | 6 | 4 |
| K | 4 | 4 | 4 | 1 |
| Others | 1 | 1 | 1 | 4 |
| Total | 80 | 78 | 103 | 92 |
| Motif | Width | Multilevel Consensus Sequence |
|---|---|---|
| 1 | 29 | EPEVTVVEJSPDDEFLILASDGLWDVLSN |
| 2 | 15 | LYVANVGDSRAVLSR |
| 3 | 11 | SFFGVFDGHGG |
| 4 | 15 | GGLAVSRAIGDRYLK |
| 5 | 29 | GGKAVQLSVDHKPBRPDERERIEAAGGRV |
| 6 | 50 | PRGGIARRLVKAALQEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVVFL |
| 7 | 15 | RGSKDBITVIVVDLK |
| 8 | 41 | DAJRKAFEATEEGFLSLVEKEWSLKPZJASVGSCCLVGVIC |
| 9 | 41 | VAEQLSAEHNASYEEVRQELQSSHPDDPHIVVLKHNVWRVK |
| 10 | 50 | HRGKGSDAAGRQDGLLWYKDLGQHVAGEFSMAVVQANQLLEDQSQVESGP |
| 11 | 41 | EDWLAALPRALVAGFVKTDKDFQTKAETSGTTVTFVIIDGW |
| 12 | 41 | SADHRLDANEEEVERVTASGGEVGRLNVVGGAEIGPLRCWP |
| 13 | 41 | AAEYLKEHLFENJLKHPEFITDTKLAISETYQKTDSEFLEA |
| 14 | 27 | FGLSSVRGRRAEMEDAVAVRPDFDDGT |
| 15 | 15 | TSGSTAVTAVVVGGH |
| 16 | 41 | KPGZGISVHEGSSKSSKLRPWGGPFLCSSCQEKKEAMEGKR |
| 17 | 37 | EEIFEEGSAMLSRRLNSNYPVRNMFKLFRCAICQVDL |
| 18 | 48 | KEVVDIVSSAPSRASAARALVESAVRAWRTKYPTSKVDDCAVVCLFLB |
| 19 | 15 | PEAADFLREHLYNNV |
| 20 | 29 | AEMAAAWREAFERAFARMDEELKGQAGVD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Zhu, L.; Cai, G.; Lv, C.; Yang, H.; Ren, X.; Hu, B.; Zhou, X.; Jiang, T.; Xiang, Y.; et al. Genome-Wide Identification and Characterization of the PP2C Family from Zea mays and Its Role in Long-Distance Signaling. Plants 2023, 12, 3153. https://doi.org/10.3390/plants12173153
Wu H, Zhu L, Cai G, Lv C, Yang H, Ren X, Hu B, Zhou X, Jiang T, Xiang Y, et al. Genome-Wide Identification and Characterization of the PP2C Family from Zea mays and Its Role in Long-Distance Signaling. Plants. 2023; 12(17):3153. https://doi.org/10.3390/plants12173153
Chicago/Turabian StyleWu, Huan, Ling Zhu, Guiping Cai, Chenxi Lv, Huan Yang, Xiaoli Ren, Bo Hu, Xuemei Zhou, Tingting Jiang, Yong Xiang, and et al. 2023. "Genome-Wide Identification and Characterization of the PP2C Family from Zea mays and Its Role in Long-Distance Signaling" Plants 12, no. 17: 3153. https://doi.org/10.3390/plants12173153
APA StyleWu, H., Zhu, L., Cai, G., Lv, C., Yang, H., Ren, X., Hu, B., Zhou, X., Jiang, T., Xiang, Y., Wei, R., Li, L., Liu, H., Muhammad, I., Xia, C., & Lan, H. (2023). Genome-Wide Identification and Characterization of the PP2C Family from Zea mays and Its Role in Long-Distance Signaling. Plants, 12(17), 3153. https://doi.org/10.3390/plants12173153







