Recent Updates on ALMT Transporters’ Physiology, Regulation, and Molecular Evolution in Plants
Abstract
:1. Introduction
2. Bioinformatic Analysis of ALMT Family Proteins
2.1. Malate and Al Recognition and Transport
2.2. ALMT/GABA Interplay
2.3. CaM Binding by ALMTs
2.4. ALMT Phosphorylation
3. Molecular Evolution
3.1. Sequence Identification
3.2. Phylogenetic Analysis
4. ALMTs in the Regulation of Stomata/Guard Cells
5. Transcriptional Regulation
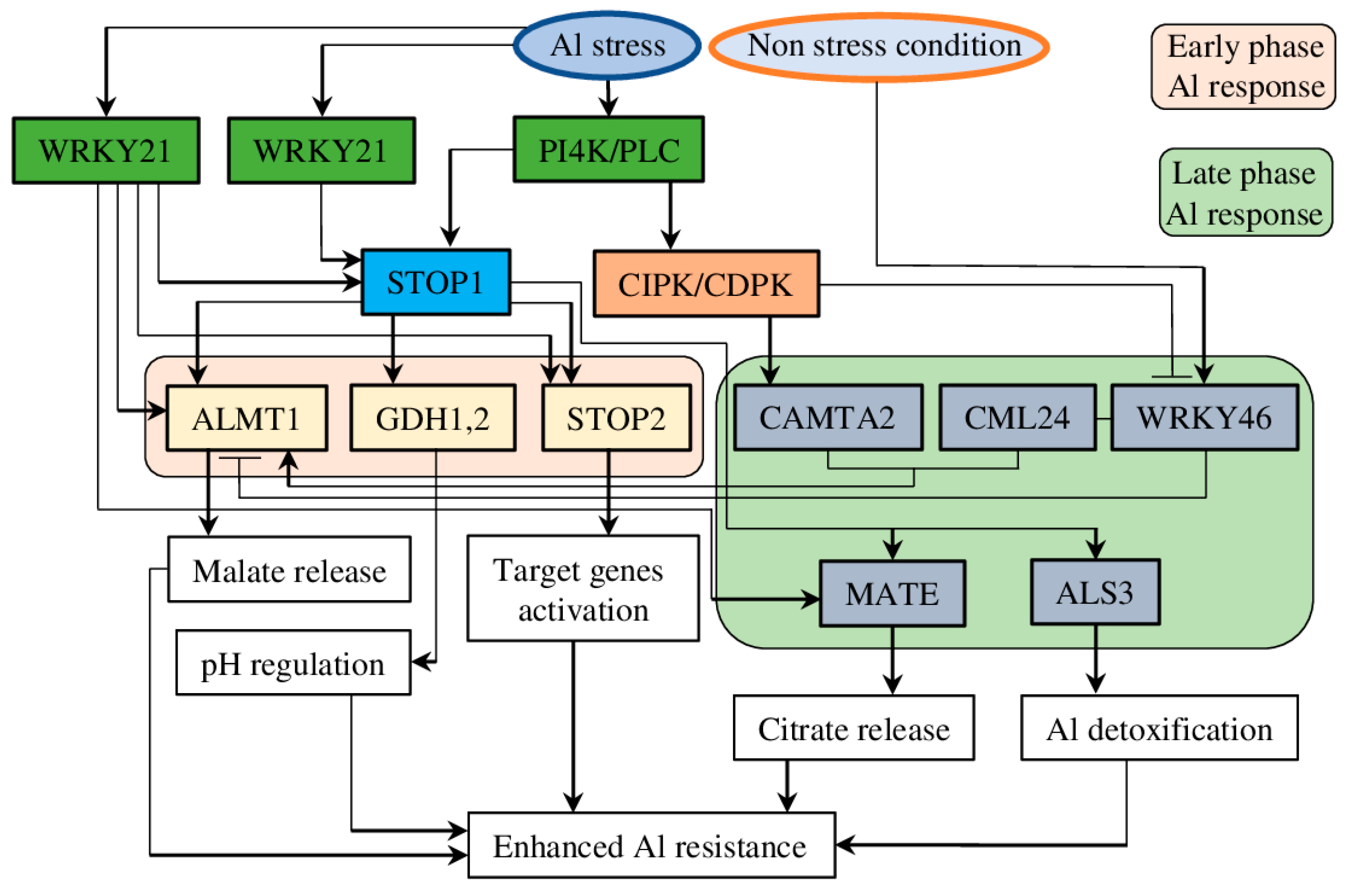
5.1. STOP1/CAMTA2/WRKY Regulatory Node
5.2. STOP1/STOP2 Pathway
5.3. STOP1/Phosphate Interplay
5.4. TFs Regulating ALMTs in Berries/Fruits
5.5. Regulation of STOP1 Proteasomal Degradation
5.6. The Role of Jasmonic Acid in ALMT Regulation
6. Conclusions and Future Prospects
- The functional role of ALMT11 is still unknown. While it misses most conserved residues required for Al/malate/GABA recognition and transport, the presented sequence similarity (especially in the TM1 region) may be sufficient to form heterodimers with other ALMTs and, thus, deactivate them to act as a negative regulator.
- Currently, the role of ALMT14 isoforms 2 and 3 is unknown. Therefore, any research investigating the functions of these proteins would be beneficial, particularly their ability to recognise/transport Al3+ ions and/or malate and the ability to form homo/hetero dimers with other ALMTs. Recent results on full-length and truncated versions of rice (Oryza sativa L.) and wheat ALMTs suggest that they function as multimeric proteins, where combinations of ALMT subunits can affect channel function [85]. Similar experiments on ALMT11 and ALMT14 would greatly advance our understanding of ALMT functionality.
- The deletion in the CTD has been identified in several ALMTs. It would be interesting to identify how this deletion affects Al3+/malate recognition/transport function, or how it correlates with protein localisation and stability.
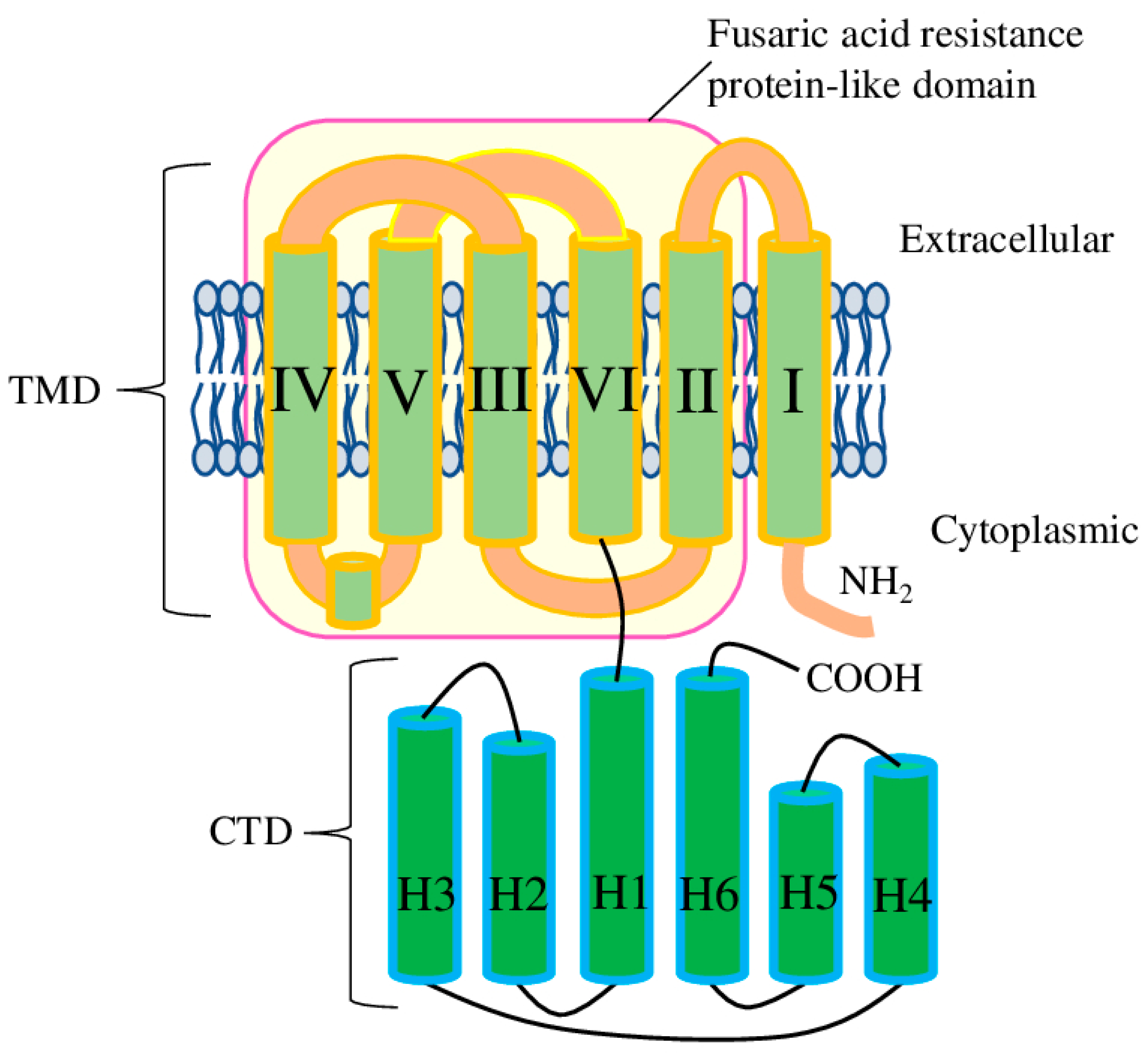
- The presence of the fusaric acid resistance protein-like (pfam13515) domain as the core of the TMD suggests that it may be the original form of the ALMT protein (before it acquired the CTD). So far, the ability of other Arabidopsis proteins possessing the fusaric acid resistance protein-like (pfam13515) domain to recognise/transport Al3+ or malate has not been studied.
- We have discussed several plant hormones interacting and regulating the STOP1/ALMT pathway of Al stress and lPi response (GABA, JA, and ABA). However, analysis of the ALMT interactome suggests that other hormones may be involved. For example, ALMT1 was shown to interact with Arabidopsis histidine kinase 4, a cytokinin receptor [86], thus suggesting possible regulation by the cytokinin.
- Also, ALMT1 interacts with a cell wall-associated receptor-like protein kinase (WAK1) [87], which is known as a receptor of oligogalacturonides, is involved in wounding response, and is a regulator of cell wall synthesis.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, Y.; Kobayashi, Y.; Sugimoto, M.; Lakshmanan, V.; Iuchi, S.; Kobayashi, M.; Bais, H.P.; Koyama, H. Characterization of the Complex Regulation of AtALMT1 Expression in Response to Phytohormones and Other Inducers. Plant Physiol. 2013, 162, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kobayashi, Y.; Watanabe, T.; Shaff, J.E.; Ohta, H.; Kochian, L.V.; Wagatsuma, T.; Kinraide, T.B.; Koyama, H. Molecular and Physiological Analysis of Al3+ and H+ Rhizotoxicities at Moderately Acidic Conditions. Plant Physiol. 2013, 163, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Tyerman, S.D.; Xu, B.; Bose, J.; Kaur, S.; Conn, V.; Domingos, P.; Ullah, S.; Wege, S.; Shabala, S.; et al. GABA Signalling Modulates Plant Growth by Directly Regulating the Activity of Plant-Specific Anion Transporters. Nat. Commun. 2015, 6, 7879. [Google Scholar] [CrossRef]
- Liang, C.; Piñeros, M.A.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, Aluminum, and Phosphorus Coordinately Regulate Malate Exudation through GmALMT1 to Improve Soybean Adaptation to Acid Soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Zhang, X.; Ramesh, S.; Gilliham, M.; Tyerman, S.D.; Zhang, W.-H. Ethylene Negatively Regulates Aluminium-Induced Malate Efflux from Wheat Roots and Tobacco Cells Transformed with TaALMT1. J. Exp. Bot. 2014, 65, 2415–2426. [Google Scholar] [CrossRef]
- Maruyama, H.; Sasaki, T.; Yamamoto, Y.; Wasaki, J. AtALMT3 Is Involved in Malate Efflux Induced by Phosphorus Deficiency in Arabidopsis thaliana Root Hairs. Plant Cell Physiol. 2019, 60, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Neuhäuser, B.; Neumann, G.; Ludewig, U. LaALMT1 Mediates Malate Release from Phosphorus-Deficient White Lupin Root Tips and Metal Root to Shoot Translocation. Plant Cell Environ. 2020, 43, 1691–1706. [Google Scholar] [CrossRef]
- Fan, W.; Xu, J.M.; Wu, P.; Yang, Z.X.; Lou, H.Q.; Chen, W.W.; Jin, J.F.; Zheng, S.J.; Yang, J.L. Alleviation by Abscisic Acid of Al Toxicity in Rice Bean Is Not Associated with Citrate Efflux but Depends on ABI5-Mediated Signal Transduction Pathways: Cross-Talk between Al and ABA Signals. J. Integr. Plant Biol. 2019, 61, 140–154. [Google Scholar] [CrossRef]
- Guo, S.-H.; Jiang, L.-Y.; Xu, Z.-M.; Li, Q.-S.; Wang, J.-F.; Ye, H.-J.; Wang, L.-L.; He, B.-Y.; Zhou, C.; Zeng, E.Y. Biological Mechanisms of Cadmium Accumulation in Edible Amaranth (Amaranthus mangostanus L.) Cultivars Promoted by Salinity: A Transcriptome Analysis. Environ. Pollut. 2020, 262, 114304. [Google Scholar] [CrossRef]
- Zarei, A.; Chiu, G.Z.; Yu, G.; Trobacher, C.P.; Shelp, B.J. Salinity-Regulated Expression of Genes Involved in GABA Metabolism and Signaling. Botany 2017, 95, 621–627. [Google Scholar] [CrossRef]
- Pereira, P.N.; Gaspar, M.; Smith, J.A.C.; Mercier, H. Ammonium Intensifies CAM Photosynthesis and Counteracts Drought Effects by Increasing Malate Transport and Antioxidant Capacity in Guzmania Monostachia. J. Exp. Bot. 2018, 69, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Ramesh, S.A.; Gilliham, M.; Tyerman, S.D.; Bose, J. Role of TaALMT1 malate-GABA Transporter in Alkaline pH Tolerance of Wheat. Plant Cell Environ. 2020, 43, 2443–2459. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, X.; Ding, Z.J.; Zhang, X.; Luo, Y.; Xu, X.; Xie, Y.; Li, X.; Yuan, T.; Zheng, S.J.; et al. Structural Basis of ALMT1-Mediated Aluminum Resistance in Arabidopsis. Cell Res. 2022, 32, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Tang, L.; Xu, J.; Zhang, X.; Zhu, Y.; Zhang, C.; Wang, M.; Liu, X.; Li, F.; Sun, F.; et al. Cryo-EM Structure and Electrophysiological Characterization of ALMT from Glycine max Reveal a Previously Uncharacterized Class of Anion Channels. Sci. Adv. 2022, 8, eabm3238. [Google Scholar] [CrossRef]
- Mumm, P.; Imes, D.; Martinoia, E.; Al-Rasheid, K.A.S.; Geiger, D.; Marten, I.; Hedrich, R. C-Terminus-Mediated Voltage Gating of Arabidopsis Guard Cell Anion Channel QUAC1. Mol. Plant 2013, 6, 1550–1563. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Piñeros, M. Functional, Structural and Phylogenetic Analysis of Domains Underlying the Al Sensitivity of the Aluminum-Activated Malate/Anion Transporter, TaALMT1. Plant J. 2013, 76, 766–780. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Kamran, M.; Sullivan, W.; Chirkova, L.; Okamoto, M.; Degryse, F.; McLaughlin, M.; Gilliham, M.; Tyerman, S.D. Aluminum-Activated Malate Transporters Can Facilitate GABA Transport. Plant Cell 2018, 30, 1147–1164. [Google Scholar] [CrossRef]
- Jaślan, J.; De Angeli, A. Heterologous Expression Reveals That GABA Does Not Directly Inhibit the Vacuolar Anion Channel At ALMT9. Plant Physiol. 2022, 189, 469–472. [Google Scholar] [CrossRef]
- Luu, K.; Rajagopalan, N.; Ching, J.C.H.; Loewen, M.C.; Loewen, M.E. The Malate-Activated ALMT12 Anion Channel in the Grass Brachypodium distachyon Is Co-Activated by Ca2+/Calmodulin. J. Biol. Chem. 2019, 294, 6142–6156. [Google Scholar] [CrossRef]
- Ligaba, A.; Kochian, L.; Piñeros, M. Phosphorylation at S384 Regulates the Activity of the TaALMT1 Malate Transporter That Underlies Aluminum Resistance in Wheat. Plant J. 2009, 60, 411–423. [Google Scholar] [CrossRef]
- Eisenach, C.; Baetz, U.; Huck, N.V.; Zhang, J.; De Angeli, A.; Beckers, G.J.M.; Martinoia, E. ABA-Induced Stomatal Closure Involves ALMT4, a Phosphorylation-Dependent Vacuolar Anion Channel of Arabidopsis. Plant Cell 2017, 29, 2552–2569. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Schäffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain Enhanced Lookup Time Accelerated BLAST. Biol. Direct 2012, 7, 12. [Google Scholar] [CrossRef]
- Sharma, T.; Dreyer, I.; Kochian, L.; Piñeros, M.A. The ALMT Family of Organic Acid Transporters in Plants and Their Involvement in Detoxification and Nutrient Security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef]
- Dreyer, I.; Gomez-Porras, J.L.; Riaño-Pachón, D.M.; Hedrich, R.; Geiger, D. Molecular Evolution of Slow and Quick Anion Channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 2012, 3, 263. [Google Scholar] [CrossRef] [PubMed]
- Daspute, A.A.; Sadhukhan, A.; Tokizawa, M.; Kobayashi, Y.; Panda, S.K.; Koyama, H. Transcriptional Regulation of Aluminum-Tolerance Genes in Higher Plants: Clarifying the Underlying Molecular Mechanisms. Front. Plant Sci. 2017, 8, 1358. [Google Scholar] [CrossRef]
- Sussmilch, F.C.; Roelfsema, M.R.G.; Hedrich, R. On the Origins of Osmotically Driven Stomatal Movements. New Phytol. 2019, 222, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.S.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 Represents an R-Type Anion Channel Required for Stomatal Movement in Arabidopsis Guard Cells: AtALMT12-Mediated Release of Anions in Guard Cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Scholz-Starke, J.; De Angeli, A.; Kovermann, P.; Burla, B.; Gambale, F.; Martinoia, E. Malate Transport by the Vacuolar AtALMT6 Channel in Guard Cells Is Subject to Multiple Regulation: AtALMT6 Mediates Malate Transport in Guard Cells. Plant J. 2011, 67, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Kovermann, P.; Meyer, S.; Hörtensteiner, S.; Picco, C.; Scholz-Starke, J.; Ravera, S.; Lee, Y.; Martinoia, E. The Arabidopsis Vacuolar Malate Channel Is a Member of the ALMT Family. Plant J. 2007, 52, 1169–1180. [Google Scholar] [CrossRef]
- De Angeli, A.; Zhang, J.; Meyer, S.; Martinoia, E. AtALMT9 Is a Malate-Activated Vacuolar Chloride Channel Required for Stomatal Opening in Arabidopsis. Nat. Commun. 2013, 4, 1804. [Google Scholar] [CrossRef]
- Sasaki, T.; Mori, I.C.; Furuichi, T.; Munemasa, S.; Toyooka, K.; Matsuoka, K.; Murata, Y.; Yamamoto, Y. Closing Plant Stomata Requires a Homolog of an Aluminum-Activated Malate Transporter. Plant Cell Physiol. 2010, 51, 354–365. [Google Scholar] [CrossRef]
- Ye, W.; Koya, S.; Hayashi, Y.; Jiang, H.; Oishi, T.; Kato, K.; Fukatsu, K.; Kinoshita, T. Identification of Genes Preferentially Expressed in Stomatal Guard Cells of Arabidopsis Thaliana and Involvement of the Aluminum-Activated Malate Transporter 6 Vacuolar Malate Channel in Stomatal Opening. Front. Plant Sci. 2021, 12, 744991. [Google Scholar] [CrossRef]
- Zhang, J.; Martinoia, E.; De Angeli, A. Cytosolic Nucleotides Block and Regulate the Arabidopsis Vacuolar Anion Channel AtALMT9. J. Biol. Chem. 2014, 289, 25581–25589. [Google Scholar] [CrossRef]
- Zhang, J.; Baetz, U.; Krügel, U.; Martinoia, E.; De Angeli, A. Identification of a Probable Pore-Forming Domain in the Multimeric Vacuolar Anion Channel AtALMT9. Plant Physiol. 2013, 163, 830–843. [Google Scholar] [CrossRef]
- Baetz, U.; Eisenach, C.; Tohge, T.; Martinoia, E.; De Angeli, A. Vacuolar Chloride Fluxes Impact Ion Content and Distribution during Early Salinity Stress. Plant Physiol. 2016, 172, 1167–1181. [Google Scholar] [CrossRef]
- Medeiros, D.B.; Martins, S.C.V.; Cavalcanti, J.H.F.; Daloso, D.M.; Martinoia, E.; Nunes-Nesi, A.; DaMatta, F.M.; Fernie, A.R.; Araújo, W.L. Enhanced Photosynthesis and Growth in Atquac1 Knockout Mutants Are Due to Altered Organic Acid Accumulation and an Increase in Both Stomatal and Mesophyll Conductance. Plant Physiol. 2016, 170, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Imes, D.; Mumm, P.; Böhm, J.; Al-Rasheid, K.A.S.; Marten, I.; Geiger, D.; Hedrich, R. Open Stomata 1 (OST1) Kinase Controls R-Type Anion Channel QUAC1 in Arabidopsis Guard Cells. Plant J. 2013, 74, 372–382. [Google Scholar] [CrossRef]
- Malcheska, F.; Ahmad, A.; Batool, S.; Müller, H.M.; Ludwig-Müller, J.; Kreuzwieser, J.; Randewig, D.; Hänsch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P.; Dias, P.N.; Tavares, B.; Portes, M.T.; Wudick, M.M.; Konrad, K.R.; Gilliham, M.; Bicho, A.; Feijó, J.A. Molecular and Electrophysiological Characterization of Anion Transport in Arabidopsis Thaliana Pollen Reveals Regulatory Roles for pH, Ca 2+ and GABA. New Phytol. 2019, 223, 1353–1371. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Tyerman, S.D.; Gilliham, M. Cytosolic GABA Inhibits Anion Transport by Wheat ALMT1. New Phytol. 2020, 225, 671–678. [Google Scholar] [CrossRef]
- Xu, B.; Long, Y.; Feng, X.; Zhu, X.; Sai, N.; Chirkova, L.; Betts, A.; Herrmann, J.; Edwards, E.J.; Okamoto, M.; et al. GABA Signalling Modulates Stomatal Opening to Enhance Plant Water Use Efficiency and Drought Resilience. Nat. Commun. 2021, 12, 1952. [Google Scholar] [CrossRef]
- Jaślan, J.; Marten, I.; Jakobson, L.; Arjus, T.; Deeken, R.; Sarmiento, C.; De Angeli, A.; Brosché, M.; Kollist, H.; Hedrich, R. ALMT-independent Guard Cell R-type Anion Currents. New Phytol. 2023, 239, 2225–2234. [Google Scholar] [CrossRef]
- Wege, S.; De Angeli, A.; Droillard, M.-J.; Kroniewicz, L.; Merlot, S.; Cornu, D.; Gambale, F.; Martinoia, E.; Barbier-Brygoo, H.; Thomine, S.; et al. Phosphorylation of the Vacuolar Anion Exchanger AtCLCa Is Required for the Stomatal Response to Abscisic Acid. Sci. Signal. 2014, 7, ra65. [Google Scholar] [CrossRef]
- Eisenach, C.; De Angeli, A. Ion Transport at the Vacuole during Stomatal Movements. Plant Physiol. 2017, 174, 520–530. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc Finger Protein STOP1 Is Critical for Proton Tolerance in Arabidopsis and Coregulates a Key Gene in Aluminum Tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, Y.; Koyama, H.; Kobayashi, M. STOP1, a Cys2/His2 Type Zinc-Finger Protein, Plays Critical Role in Acid Soil Tolerance in Arabidopsis. Plant Signal. Behav. 2008, 3, 128–130. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Li, G.X.; Zheng, S.J. WRKY46 Functions as a Transcriptional Repressor of ALMT1, Regulating Aluminum-Induced Malate Secretion in Arabidopsis. Plant J. 2013, 76, 825–835. [Google Scholar] [CrossRef]
- Tokizawa, M.; Kobayashi, Y.; Saito, T.; Kobayashi, M.; Iuchi, S.; Nomoto, M.; Tada, Y.; Yamamoto, Y.Y.; Koyama, H. SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and Other Transcription Factors Are Involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 Expression. Plant Physiol. 2015, 167, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Ligaba-Osena, A.; Fei, Z.; Liu, J.; Xu, Y.; Shaff, J.; Lee, S.; Luan, S.; Kudla, J.; Kochian, L.; Piñeros, M. Loss-of-function Mutation of the Calcium Sensor CBL 1 Increases Aluminum Sensitivity in Arabidopsis. New Phytol. 2017, 214, 830–841. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, P.; Bai, Z.; Herde, M.; Ma, Y.; Li, N.; Liu, S.; Huang, C.; Cui, R.; Ma, H.; et al. Calmodulin-like Protein CML24 Interacts with CAMTA2 and WRKY46 to Regulate ALMT1 -dependent Al Resistance in Arabidopsis thaliana. New Phytol. 2022, 233, 2471–2487. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Sadhukhan, A.; Kobayashi, Y.; Ogo, N.; Tokizawa, M.; Agrahari, R.K.; Ito, H.; Iuchi, S.; Kobayashi, M.; Asai, A.; et al. Involvement of Phosphatidylinositol Metabolism in Aluminum-Induced Malate Secretion in Arabidopsis. J. Exp. Bot. 2019, 70, 3329–3342. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wang, J.; Wang, X.; Zhang, X.; Cheng, Y.; Cai, Z.; Nian, H.; Ma, Q. GmWRKY21, a Soybean WRKY Transcription Factor Gene, Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 833326. [Google Scholar] [CrossRef]
- Tokizawa, M.; Enomoto, T.; Ito, H.; Wu, L.; Kobayashi, Y.; Mora-Macías, J.; Armenta-Medina, D.; Iuchi, S.; Kobayashi, M.; Nomoto, M.; et al. High Affinity Promoter Binding of STOP1 Is Essential for Early Expression of Novel Aluminum-Induced Resistance Genes GDH1 and GDH2 in Arabidopsis. J. Exp. Bot. 2021, 72, 2769–2789. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ohyama, Y.; Kobayashi, Y.; Ito, H.; Iuchi, S.; Fujita, M.; Zhao, C.-R.; Tanveer, T.; Ganesan, M.; Kobayashi, M.; et al. STOP2 Activates Transcription of Several Genes for Al- and Low PH-Tolerance That Are Regulated by STOP1 in Arabidopsis. Mol. Plant 2014, 7, 311–322. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Yang, C.; Cheng, Y.; Han, Z.; Cai, Z.; Nian, H.; Ma, Q. GsMAS1 Encoding a MADS-Box Transcription Factor Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. IJMS 2020, 21, 2004. [Google Scholar] [CrossRef]
- Balzergue, C.; Dartevelle, T.; Godon, C.; Laugier, E.; Meisrimler, C.; Teulon, J.-M.; Creff, A.; Bissler, M.; Brouchoud, C.; Hagège, A.; et al. Low Phosphate Activates STOP1-ALMT1 to Rapidly Inhibit Root Cell Elongation. Nat. Commun. 2017, 8, 15300. [Google Scholar] [CrossRef] [PubMed]
- Mora-Macías, J.; Ojeda-Rivera, J.O.; Gutiérrez-Alanís, D.; Yong-Villalobos, L.; Oropeza-Aburto, A.; Raya-González, J.; Jiménez-Domínguez, G.; Chávez-Calvillo, G.; Rellán-Álvarez, R.; Herrera-Estrella, L. Malate-Dependent Fe Accumulation Is a Critical Checkpoint in the Root Developmental Response to Low Phosphate. Proc. Natl. Acad. Sci. USA 2017, 114, E3563–E3572. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Z.; Zheng, Z.; Dong, J.; Song, L.; Sui, L.; Nussaume, L.; Desnos, T.; Liu, D. Genetic Dissection of Fe-Dependent Signaling in Root Developmental Responses to Phosphate Deficiency. Plant Physiol. 2019, 179, 300–316. [Google Scholar] [CrossRef] [PubMed]
- Godon, C.; Mercier, C.; Wang, X.; David, P.; Richaud, P.; Nussaume, L.; Liu, D.; Desnos, T. Under Phosphate Starvation Conditions, Fe and Al Trigger Accumulation of the Transcription Factor STOP1 in the Nucleus of Arabidopsis Root Cells. Plant J. 2019, 99, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, H.; Wang, N.; Fan, H.; Chen, C.; Cui, Y.; Liu, H.; Ling, H. Mediator Subunit 16 Functions in the Regulation of Iron Uptake Gene Expression in Arabidopsis. New Phytol. 2014, 203, 770–783. [Google Scholar] [CrossRef]
- Raya-González, J.; Ojeda-Rivera, J.O.; Mora-Macias, J.; Oropeza-Aburto, A.; Ruiz-Herrera, L.F.; López-Bucio, J.; Herrera-Estrella, L. MEDIATOR16 Orchestrates Local and Systemic Responses to Phosphate Scarcity in Arabidopsis Roots. New Phytol. 2021, 229, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Hou, S.; Tu, G.; Lan, W.; Jing, Y. Transcription Factor WRKY33 Mediates the Phosphate Deficiency-Induced Remodeling of Root Architecture by Modulating Iron Homeostasis in Arabidopsis Roots. IJMS 2021, 22, 9275. [Google Scholar] [CrossRef]
- Lee, H.-S.; Kim, Y.; Pham, G.; Kim, J.W.; Song, J.-H.; Lee, Y.; Hwang, Y.-S.; Roux, S.J.; Kim, S.-H. Brassinazole Resistant 1 (BZR1)-Dependent Brassinosteroid Signalling Pathway Leads to Ectopic Activation of Quiescent Cell Division and Suppresses Columella Stem Cell Differentiation. EXBOTJ 2015, 66, 4835–4849. [Google Scholar] [CrossRef]
- Liu, T.; Deng, S.; Zhang, C.; Yang, X.; Shi, L.; Xu, F.; Wang, S.; Wang, C. Brassinosteroid Signaling Regulates Phosphate Starvation-induced Malate Secretion in Plants. JIPB 2023, 65, 1099–1112. [Google Scholar] [CrossRef]
- Xu, J.M.; Wang, Z.Q.; Wang, J.Y.; Li, P.F.; Jin, J.F.; Chen, W.W.; Fan, W.; Kochian, L.V.; Zheng, S.J.; Yang, J.L. Low Phosphate Represses Histone Deacetylase Complex1 to Regulate Root System Architecture Remodeling in Arabidopsis. New Phytol. 2020, 225, 1732–1745. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Wang, C.-K.; Zhao, Y.-W.; Sun, C.-H.; Hu, D.-G. Mechanisms and Regulation of Organic Acid Accumulation in Plant Vacuoles. Hortic. Res. 2021, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.-G.; Li, Y.-Y.; Zhang, Q.-Y.; Li, M.; Sun, C.-H.; Yu, J.-Q.; Hao, Y.-J. The R2R3-MYB Transcription Factor MdMYB73 Is Involved in Malate Accumulation and Vacuolar Acidification in Apple. Plant J. 2017, 91, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Gu, K.-D.; Cheng, L.; Wang, J.-H.; Yu, J.-Q.; Wang, X.-F.; You, C.-X.; Hu, D.-G.; Hao, Y.-J. BTB-TAZ Domain Protein MdBT2 Modulates Malate Accumulation and Vacuolar Acidification in Response to Nitrate. Plant Physiol. 2020, 183, 750–764. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Gu, K.-D.; Wang, J.-H.; Yu, J.-Q.; Wang, X.-F.; Zhang, S.; You, C.-X.; Hu, D.-G.; Hao, Y.-J. BTB-BACK-TAZ Domain Protein MdBT2-Mediated MdMYB73 Ubiquitination Negatively Regulates Malate Accumulation and Vacuolar Acidification in Apple. Hortic. Res. 2020, 7, 151. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.-F. F-Box Protein RAE1 Regulates the Stability of the Aluminum-Resistance Transcription Factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhou, F.; Zhang, Y.; Singh, S.; Huang, C. Degradation of STOP1 Mediated by the F-box Proteins RAH1 and RAE1 Balances Aluminum Resistance and Plant Growth in Arabidopsis thaliana. Plant J. 2021, 106, 493–506. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.; Huang, C. Mutation of HPR1 Encoding a Component of the THO/TREX Complex Reduces STOP1 Accumulation and Aluminium Resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193. [Google Scholar] [CrossRef]
- Luna, R.; Rondón, A.G.; Aguilera, A. New Clues to Understand the Role of THO and Other Functionally Related Factors in MRNP Biogenesis. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2012, 1819, 514–520. [Google Scholar] [CrossRef]
- Zhu, Y.-F.; Guo, J.; Zhang, Y.; Huang, C.-F. The THO/TREX Complex Component RAE2/TEX1 Is Involved in the Regulation of Aluminum Resistance and Low Phosphate Response in Arabidopsis. Front. Plant Sci. 2021, 12, 698443. [Google Scholar] [CrossRef]
- Augustine, R.C.; Vierstra, R.D. SUMOylation: Re-Wiring the Plant Nucleus during Stress and Development. Curr. Opin. Plant Biol. 2018, 45, 143–154. [Google Scholar] [CrossRef]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; Van Den Burg, H.A.; Huang, C.-F. Regulation of Aluminum Resistance in Arabidopsis Involves the SUMOylation of the Zinc Finger Transcription Factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhang, J.; Yang, D.-L.; Huang, C.-F. The SUMO E3 Ligase SIZ1 Partially Regulates STOP1 SUMOylation and Stability in Arabidopsis thaliana. Plant Signal. Behav. 2021, 16, 1899487. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-B.; He, C.; Ma, Y.; Herde, M.; Ding, Z. Jasmonic Acid Enhances Al-Induced Root Growth Inhibition. Plant Physiol. 2017, 173, 1420–1433. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Su, H.; Guo, L.; Zhang, J.; Li, Y.; Xu, J.; Zhang, X.; Guo, Y.-D.; Zhang, N. Jasmonate and Aluminum Crosstalk in Tomato: Identification and Expression Analysis of WRKYs and ALMTs during JA/Al-Regulated Root Growth. Plant Physiol. Biochem. 2020, 154, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Hu, Z.; Luo, Y.; Feng, C.; Long, Y. Multiple ALMT Subunits Combine to Form Functional Anion Channels: A Case Study for Rice ALMT7. Front. Plant Sci. 2022, 13, 1012578. [Google Scholar] [CrossRef] [PubMed]
- Dortay, H.; Gruhn, N.; Pfeifer, A.; Schwerdtner, M.; Schmülling, T.; Heyl, A. Toward an Interaction Map of the Two-Component Signaling Pathway of Arabidopsis thaliana. J. Proteome Res. 2008, 7, 3649–3660. [Google Scholar] [CrossRef]
- Jones, A.M.; Xuan, Y.; Xu, M.; Wang, R.-S.; Ho, C.-H.; Lalonde, S.; You, C.H.; Sardi, M.I.; Parsa, S.A.; Smith-Valle, E.; et al. Border Control—A Membrane-Linked Interactome of Arabidopsis. Science 2014, 344, 711–716. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabravolski, S.A.; Isayenkov, S.V. Recent Updates on ALMT Transporters’ Physiology, Regulation, and Molecular Evolution in Plants. Plants 2023, 12, 3167. https://doi.org/10.3390/plants12173167
Dabravolski SA, Isayenkov SV. Recent Updates on ALMT Transporters’ Physiology, Regulation, and Molecular Evolution in Plants. Plants. 2023; 12(17):3167. https://doi.org/10.3390/plants12173167
Chicago/Turabian StyleDabravolski, Siarhei A., and Stanislav V. Isayenkov. 2023. "Recent Updates on ALMT Transporters’ Physiology, Regulation, and Molecular Evolution in Plants" Plants 12, no. 17: 3167. https://doi.org/10.3390/plants12173167
APA StyleDabravolski, S. A., & Isayenkov, S. V. (2023). Recent Updates on ALMT Transporters’ Physiology, Regulation, and Molecular Evolution in Plants. Plants, 12(17), 3167. https://doi.org/10.3390/plants12173167






