Genome-Wide Identification and Expression Analysis of RLCK-VII Subfamily Genes Reveal Their Roles in Stress Responses of Upland Cotton
Abstract
:1. Introduction
2. Result
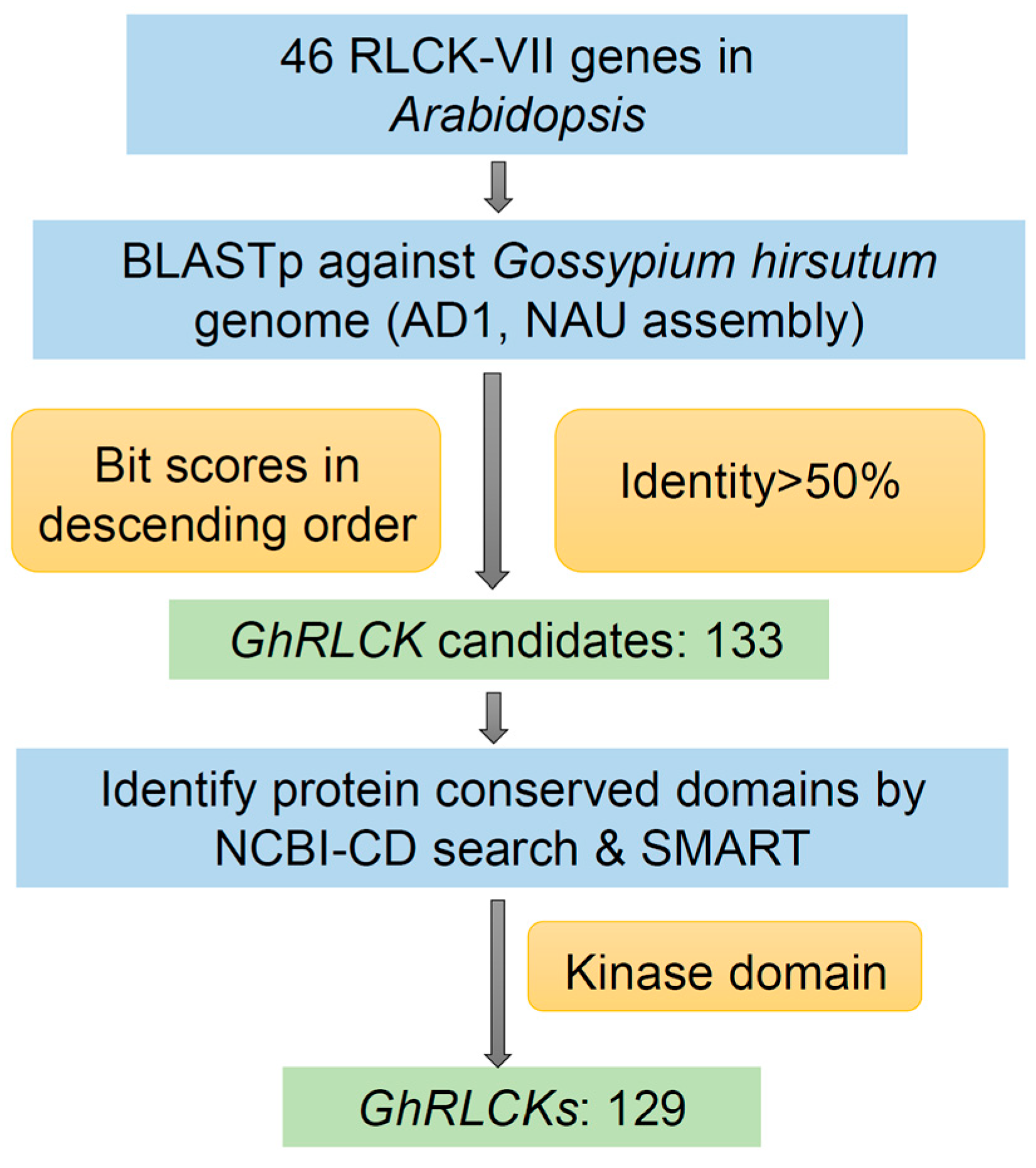
2.1. Identification of RLCK-VII Subfamily Genes in Gossypium hirsutum
2.2. Phylogenetic Analysis of RLCK VII Subfamily Genes in Cotton and Arabidopsis
2.3. Conserved Domains and Structure Analysis of RLCK-VII Genes in G. hirsutum and A. thaliana
2.4. Chromosomal Location, Gene Duplication, and Synteny Analysis of GhRLCKs
2.5. Tissue- and Organ-Specific Expression Profiling of GhRLCKs in Upland Cotton
2.6. Expression Patterns of GhRLCK Genes in Upland Cotton under Abiotic Stresses
2.7. Expression Patterns of GhRLCKs under V. dahliae Infection
2.8. Cis-Elements in GhRLCK Promoters
2.9. Silencing of GhRLCK7 Compromised Resistance to V. dahliae in Upland Cotton
3. Discussion
3.1. The Evolutionary Patterns of GhRLCKs
3.2. Potential Functions of GhRLCK Genes in Cotton’s Response to Abiotic Stresses
3.3. Involvement of GhRLCKs in Cotton Defense against V. dahliae
4. Materials and Methods
4.1. Identification of RLCK-VII Subfamily Genes in Upland Cotton
4.2. Physicochemical Property Characterization of GhRLCK Proteins
4.3. Phylogenetic Analysis of GhRLCK Genes
4.4. Gene Structure and Conserved Domain Analysis
4.5. Chromosomal Location, Gene Duplication, and Synteny Analysis
4.6. Gene Expression Profile Analysis
4.7. Analysis of Promoter Regions for Cis-Elements
4.8. Cultivation of Cotton and V. dahliae
4.9. Treatments of Cotton with Abiotic and Biotic Stresses
4.10. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
4.11. Construction of VIGS Vector and Implementation of VIGS
4.12. Disease Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.B.; Li, Y.; Wang, B.; Chee, P.W. Recent advances in cotton genomics. Int. J. Plant Genom. 2008, 2008, 742304. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wendel, J.F.; Hua, J. Designations for individual genomes and chromosomes in Gossypium. J. Cotton Res. 2018, 1, 3. [Google Scholar] [CrossRef]
- Chen, Z.J.; Scheffler, B.E.; Dennis, E.; Triplett, B.A.; Zhang, T.; Guo, W.; Chen, X.; Stelly, D.M.; Rabinowicz, P.D.; Town, C.D.; et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F. New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad. Sci. USA 1989, 86, 4132–4136. [Google Scholar] [CrossRef]
- Wang, K.B.; Wang, Z.W.; Li, F.G.; Ye, W.W.; Wang, J.Y.; Song, G.L.; Yue, Z.; Cong, L.; Shang, H.H.; Zhu, S.L.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Li, F.G.; Fan, G.Y.; Wang, K.B.; Sun, F.M.; Yuan, Y.L.; Song, G.L.; Li, Q.; Ma, Z.Y.; Lu, C.R.; Zou, C.S.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef]
- Li, F.G.; Fan, G.Y.; Lu, C.R.; Xiao, G.H.; Zou, C.S.; Kohel, R.J.; Ma, Z.Y.; Shang, H.H.; Ma, X.F.; Wu, J.Y.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Zhang, T.Z.; Hu, Y.; Jiang, W.K.; Fang, L.; Guan, X.Y.; Chen, J.D.; Zhang, J.B.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Yuan, D.J.; Tang, Z.H.; Wang, M.J.; Gao, W.H.; Tu, L.L.; Jin, X.; Chen, L.L.; He, Y.H.; Zhang, L.; Zhu, L.F.; et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2015, 5, 17662. [Google Scholar] [CrossRef]
- Saranga, Y.; Paterson, A.H.; Levi, A. Bridging Classical and Molecular Genetics of Abiotic Stress Resistance in Cotton. In Genetics and Genomics of Cotton; Paterson, A.H., Ed.; Springer: New York, NY, USA, 2009; pp. 337–352. [Google Scholar]
- Billah, M.; Li, F.; Yang, Z. Regulatory Network of Cotton Genes in Response to Salt, Drought and Wilt Diseases (Verticillium and Fusarium): Progress and Perspective. Front. Plant Sci. 2021, 12, 759245. [Google Scholar] [CrossRef] [PubMed]
- Ranga, A.; Kak, V.; Darvhankar, M. Genetic and molecular research of resistance to wilt in cotton: A concise review. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 2410–2422. [Google Scholar] [CrossRef]
- Wagner, T.A.; Gu, A.; Duke, S.E.; Bell, A.A.; Magill, C.; Liu, J. Genetic Diversity and Pathogenicity of Verticillium dahliae Isolates and Their Co-occurrence with Fusarium oxysporum f. sp. vasinfectum Causing Cotton Wilt in Xinjiang, China. Plant Dis. 2021, 105, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Qanmber, G.; Wang, Z.; Yang, Z.; Li, F. Gossypium Genomics: Trends, Scope, and Utilization for Cotton Improvement. Trends Plant Sci. 2020, 25, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Ma, X.; Shan, L.; He, P. Big roles of small kinases: The complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol. 2013, 55, 1188–1197. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.S.; Tzeng, Y.H.; Mayer, K.F.X.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Rao, S.; Zhou, Z.; Miao, P.; Bi, G.; Hu, M.; Wu, Y.; Feng, F.; Zhang, X.; Zhou, J.M. Roles of Receptor-Like Cytoplasmic Kinase VII Members in Pattern-Triggered Immune Signaling. Plant Physiol. 2018, 177, 1679–1690. [Google Scholar] [CrossRef]
- Sreekanta, S.; Bethke, G.; Hatsugai, N.; Tsuda, K.; Thao, A.; Wang, L.; Katagiri, F.; Glazebrook, J. The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol. 2015, 207, 78–90. [Google Scholar] [CrossRef]
- Kong, Q.; Sun, T.J.; Qu, N.; Ma, J.L.; Li, M.; Cheng, Y.T.; Zhang, Q.; Wu, D.; Zhang, Z.B.; Zhang, Y.L. Two Redundant Receptor-Like Cytoplasmic Kinases Function Downstream of Pattern Recognition Receptors to Regulate Activation of SA Biosynthesis. Plant Physiol. 2016, 171, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014, 79, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Xue, J.; Wang, F.Z.; Huang, X.J.; Gong, B.Q.; Tao, Y.H.; Shen, W.Z.; Tao, K.H.; Yao, N.; Xiao, S.; et al. Plasma membrane-nucleo-cytoplasmic coordination of a receptor-like cytoplasmic kinase promotes EDS1-dependent plant immunity. Nat. Plants 2022, 8, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Du, C.Q.; Li, X.S.; Chen, J.; Chen, W.J.; Li, B.; Li, C.Y.; Wang, L.; Li, J.L.; Zhao, X.Y.; Lin, J.Z.; et al. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8326–E8334. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J. 2011, 66, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Ao, Y.; Feng, D.R.; Liu, J.; Wang, J.F.; Wang, H.B.; Liu, B. OsRLCK 57, OsRLCK107 and OsRLCK118 Positively Regulate Chitin- and PGN-Induced Immunity in Rice. Rice 2017, 10, 6. [Google Scholar] [CrossRef]
- Zhou, X.G.; Wang, J.; Peng, C.F.; Zhu, X.B.; Yin, J.J.; Li, W.T.; He, M.; Wang, J.C.; Chern, M.; Yuan, C.; et al. Four receptor-like cytoplasmic kinases regulate development and immunity in rice. Plant Cell Environ. 2016, 39, 1381–1392. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yoshimura, Y.; Nakagawa, S.; Mezaki, H.; Yoshimura, S.; Kawasaki, T. OsDRE2 contributes to chitin-triggered response through its interaction with OsRLCK185. Biosci. Biotechnol. Biochem. 2019, 83, 281–290. [Google Scholar] [CrossRef]
- Ramegowda, V.; Basu, S.; Gupta, C.; Pereira, A. Regulation of grain yield in rice under well-watered and drought stress conditions by GUDK. Plant Signal. Behav. 2015, 10, e1034421. [Google Scholar] [CrossRef]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef]
- Shen, W.; Gómez-Cadenas, A.; Routly, E.L.; Ho, T.H.; Simmonds, J.A.; Gulick, P.J. The salt stress-inducible protein kinase gene, Esi47, from the salt-tolerant wheatgrass Lophopyrum elongatum is involved in plant hormone signaling. Plant Physiol. 2001, 125, 1429–1441. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, L.; Jamieson, P.; Zhang, L.; Zhao, Z.; Babilonia, K.; Shao, W.; Wu, L.; Mustafa, R.; Amin, I.; et al. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell 2020, 32, 3978–4001. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Y.; Zhang, M.; Rong, K.; Wu, Y.; Zhang, M.; Hu, H. Genome-wide identification of xylan glucuronosyltransferase family in cotton and function characterization of GhGUX5 in regulating Verticillium wilt resistance. Int. J. Biol. Macromol. 2023, 245, 124795. [Google Scholar] [CrossRef]
- Dai, M.H.; Zhou, N.; Zhang, Y.; Zhang, Y.X.; Ni, K.S.; Wu, Z.L.; Liu, L.Y.; Wang, X.G.; Chen, Q.J. Genome-wide analysis of the SBT gene family involved in drought tolerance in cotton. Front. Plant Sci. 2023, 13, 1097732. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Cheng, Y.; Vanitha, J.; Kumar, N.; Ramamoorthy, R.; Ramachandran, S.; Jiang, S.Y. Expansion Mechanisms and Functional Divergence of the Glutathione S-Transferase Family in Sorghum and Other Higher Plants. DNA Res. 2011, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, J.; Huang, Y.; Wang, S.; Wei, L.; Liu, D.; Weng, Y.; Xiang, J.; Zhu, Q.; Yang, Z.; et al. CottonMD: A multi-omics database for cotton biological study. Nucleic Acids Res. 2023, 51, D1446–D1456. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, M.; Li, N.; Wang, H.; Qiu, P.; Pei, L.; Xu, Z.; Wang, T.; Gao, E.; Liu, J.; et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 2018, 16, 1172–1185. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Shen, J.L.; Li, W.J.; Wu, N.; Chen, C.; Hou, Y.X. Evolutionary and Characteristic Analysis of RING-DUF1117 E3 Ubiquitin Ligase Genes in Gossypium Discerning the Role of GhRDUF4D in Verticillium dahliae Resistance. Biomolecules 2021, 11, 1145. [Google Scholar] [CrossRef] [PubMed]
- Cronn, R.C.; Small, R.L.; Wendel, J.F. Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 1999, 96, 14406–14411. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, Y.Y.; Chen, D.D.; Liu, D.; Hu, M.J.; Dong, J.; Zhang, X.P.; Song, L.R.; Shen, F.F. The Catalase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef]
- Cao, J.; Shi, F. Evolution of the RALF Gene Family in Plants: Gene Duplication and Selection Patterns. Evol. Bioinform. 2012, 8, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.M.; Chang, C.R.; Schaller, G.E. Perception of Ethylene by Plants—Ethylene Receptors. In Annual Plant Reviews Volume 44: Plant Hormone Ethylene; McManus, M.T., Ed.; Wiley: Hoboken, NJ, USA, 2012; pp. 117–145. [Google Scholar]
- Zhu, Y.; Zhao, M.; Li, T.; Wang, L.; Liao, C.; Liu, D.; Zhang, H.; Zhao, Y.; Liu, L.; Ge, X.; et al. Interactions between Verticillium dahliae and cotton: Pathogenic mechanism and cotton resistance mechanism to Verticillium wilt. Front. Plant Sci. 2023, 14, 1174281. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Liu, Y.; Maierhofer, T.; Rybak, K.; Sklenar, J.; Breakspear, A.; Johnston, M.G.; Fliegmann, J.; Huang, S.; Roelfsema, M.R.G.; Felix, G.; et al. Anion channel SLAH3 is a regulatory target of chitin receptor-associated kinase PBL27 in microbial stomatal closure. Elife 2019, 8, e44474. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-Mediated Chitin Perception and Immune Signaling Requires Receptor-like Cytoplasmic Kinase 185 to Activate an MAPK Cascade in Rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S.; et al. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Wei, F.; Coe, E.; Nelson, W.; Bharti, A.K.; Engler, F.; Butler, E.; Kim, H.; Goicoechea, J.L.; Chen, M.; Lee, S.; et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007, 3, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. Cell Mol. Biol. 2011, 66, 293–305. [Google Scholar] [CrossRef] [PubMed]












| Gene Locus ID | Name | Length/bp | CDS Length/bp | Number of Exons | Number of Amino Acids | Molecular Weight /Da | Theoretical pI | Instability Index | Aliphatic Index |
|---|---|---|---|---|---|---|---|---|---|
| Gh_A01G0195 | GhRLCK8A | 3040 | 1554 | 5 | 517 | 57,041.98 | 9.09 | 37.98 | 66.63 |
| Gh_A01G0607 | GhRLCK1A | 1912 | 1176 | 5 | 391 | 43,474.88 | 4.95 | 40.14 | 75.37 |
| Gh_A01G0842 | GhRLCK22A | 3180 | 1182 | 6 | 393 | 43,634.61 | 8.83 | 37.12 | 82.62 |
| Gh_A01G1911 | GhRLCK63A | 2752 | 1116 | 6 | 371 | 41,226.76 | 9.25 | 42.22 | 79.92 |
| Gh_A01G2145 | GhRLCK11A | 3657 | 1200 | 5 | 399 | 44,214.81 | 9.53 | 34.11 | 75.81 |
| Gh_A02G0005 | GhRLCK45A | 3462 | 1281 | 6 | 426 | 47,181.73 | 8.91 | 35.75 | 83.10 |
| Gh_A02G0870 | GhRLCK59A | 3266 | 1272 | 6 | 423 | 46,399.66 | 9.29 | 26.04 | 75.15 |
| Gh_A02G1028 | GhRLCK68A | 1947 | 1437 | 6 | 478 | 52,031.13 | 6.56 | 55.10 | 86.67 |
| Gh_A02G1701 | GhRLCK58A | 7322 | 1275 | 6 | 424 | 47,405.54 | 8.79 | 36.09 | 74.95 |
| Gh_A03G0052 | GhRLCK47A | 2196 | 1002 | 5 | 333 | 36,755.70 | 9.25 | 42.84 | 77.93 |
| Gh_A03G0179 | GhRLCK46A | 5702 | 1197 | 7 | 398 | 44,844.30 | 6.28 | 38.58 | 87.71 |
| Gh_A03G1241 | GhRLCK35A | 6991 | 1047 | 7 | 348 | 39,554.32 | 9.03 | 43.49 | 85.43 |
| Gh_A03G1511 | GhRLCK55A | 2608 | 1266 | 7 | 421 | 47,057.59 | 9.67 | 36.11 | 77.58 |
| Gh_A03G2069 | GhRLCK61A | 1906 | 1287 | 6 | 428 | 47,415.63 | 9.19 | 40.87 | 80.02 |
| Gh_A04G0613 | GhRLCK31A | 2600 | 1224 | 4 | 407 | 46,151.73 | 9.60 | 41.83 | 79.48 |
| Gh_A04G1161 | GhRLCK33A | 3355 | 1080 | 7 | 359 | 40,469.48 | 9.00 | 44.45 | 83.90 |
| Gh_A05G0076 | GhRLCK40A | 1683 | 1305 | 6 | 434 | 48,511.47 | 9.51 | 38.21 | 83.57 |
| Gh_A05G0253 | GhRLCK53A | 1709 | 1356 | 5 | 451 | 51,229.67 | 9.17 | 41.72 | 79.33 |
| Gh_A05G0599 | GhRLCK66A | 2513 | 1149 | 6 | 382 | 41,979.78 | 9.44 | 29.68 | 79.42 |
| Gh_A05G0809 | GhRLCK28A | 1940 | 1401 | 4 | 466 | 52,446.62 | 8.75 | 36.72 | 83.43 |
| Gh_A05G0810 | GhRLCK60A | 2316 | 1218 | 5 | 405 | 44,786.69 | 9.58 | 36.34 | 74.96 |
| Gh_A05G1164 | GhRLCK48A | 1729 | 1344 | 5 | 477 | 50,704.40 | 9.43 | 43.57 | 85.46 |
| Gh_A05G2537 | GhRLCK29A | 2178 | 1281 | 4 | 426 | 47,965.80 | 9.49 | 43.98 | 84.44 |
| Gh_A05G2570 | GhRLCK10A | 3045 | 1152 | 5 | 383 | 42,251.36 | 9.26 | 27.27 | 81.78 |
| Gh_A05G3044 | GhRLCK64A | 2475 | 1236 | 6 | 411 | 45,474.55 | 9.45 | 42.55 | 76.72 |
| Gh_A05G3371 | GhRLCK19A | 2613 | 1557 | 5 | 518 | 57,795.10 | 7.61 | 58.61 | 61.18 |
| Gh_A05G3532 | GhRLCK42A | 4149 | 1185 | 5 | 394 | 44,234.99 | 6.35 | 36.28 | 76.24 |
| Gh_A06G0688 | GhRLCK54A | 1610 | 1332 | 4 | 443 | 49,645.87 | 8.98 | 44.06 | 80.56 |
| Gh_A06G0724 | GhRLCK50A | 1621 | 1017 | 5 | 338 | 38,136.74 | 8.03 | 30.61 | 85.38 |
| Gh_A06G1294 | GhRLCK4A | 1817 | 1170 | 6 | 389 | 43,541.75 | 6.12 | 35.59 | 85.73 |
| Gh_A07G0319 | GhRLCK41A | 2954 | 1284 | 6 | 427 | 47,613.07 | 9.23 | 32.50 | 78.99 |
| Gh_A07G2351 | GhRLCK51A | 1974 | 1152 | 7 | 383 | 43,458.78 | 7.67 | 42.42 | 85.74 |
| Gh_A08G0138 | GhRLCK44A | 5083 | 1356 | 6 | 454 | 50,793.98 | 9.22 | 37.85 | 79.19 |
| Gh_A08G0245 | GhRLCK26A | 2441 | 1251 | 4 | 416 | 46,582.46 | 9.09 | 36.01 | 82.96 |
| Gh_A08G1460 | GhRLCK23A | 1862 | 1170 | 4 | 389 | 43,793.83 | 9.51 | 40.09 | 76.89 |
| Gh_A08G2568 | GhRLCK14A | 1875 | 1269 | 5 | 422 | 46,882.09 | 8.04 | 36.85 | 75.12 |
| Gh_A09G0212 | GhRLCK36A | 3028 | 1140 | 6 | 379 | 42,155.86 | 9.35 | 52.32 | 68.97 |
| Gh_A09G0371 | GhRLCK21A | 3221 | 1284 | 6 | 427 | 47,536.09 | 8.28 | 25.85 | 77.24 |
| Gh_A09G0599 | GhRLCK6A | 4986 | 1545 | 5 | 514 | 56,646.53 | 9.10 | 34.06 | 66.42 |
| Gh_A09G1001 | GhRLCK67A | 3885 | 1236 | 6 | 411 | 45,302.56 | 9.30 | 26.20 | 77.59 |
| Gh_A09G1391 | GhRLCK32A | 2426 | 1323 | 4 | 440 | 49,995.39 | 9.47 | 47.08 | 83.50 |
| Gh_A09G1456 | GhRLCK62A | 1787 | 1194 | 6 | 397 | 44,277.17 | 9.78 | 39.91 | 76.17 |
| Gh_A09G2123 | GhRLCK5A | 2527 | 1506 | 5 | 501 | 55,546.36 | 8.80 | 36.21 | 68.94 |
| Gh_A10G0498 | GhRLCK49A | 2140 | 1404 | 6 | 467 | 52,688.41 | 9.33 | 39.45 | 88.09 |
| Gh_A10G1428 | GhRLCK7A | 4007 | 1548 | 5 | 515 | 56,653.56 | 8.99 | 38.89 | 68.02 |
| Gh_A10G1831 | GhRLCK30A | 2502 | 1290 | 4 | 429 | 49,065.08 | 9.71 | 42.53 | 82.03 |
| Gh_A10G1861 | GhRLCK9A | 2450 | 1152 | 5 | 323 | 35,908.23 | 8.68 | 31.52 | 89.72 |
| Gh_A10G1973 | GhRLCK65A | 2506 | 1161 | 6 | 386 | 42,796.62 | 9.12 | 35.70 | 76.55 |
| Gh_A11G0259 | GhRLCK25A | 1764 | 1164 | 4 | 387 | 43,458.52 | 9.65 | 35.03 | 78.32 |
| Gh_A11G0581 | GhRLCK20A | 2382 | 1242 | 5 | 413 | 45,673.01 | 8.48 | 34.63 | 73.22 |
| Gh_A11G0673 | GhRLCK27A | 2199 | 1317 | 4 | 438 | 49,421.60 | 9.35 | 39.55 | 79.50 |
| Gh_A11G1264 | GhRLCK16A | 1666 | 1290 | 5 | 429 | 47,253.97 | 5.81 | 37.23 | 73.52 |
| Gh_A11G2714 | GhRLCK17A | 3936 | 1710 | 5 | 569 | 61,686.29 | 6.08 | 48.05 | 62.46 |
| Gh_A11G3042 | GhRLCK37A | 3132 | 1431 | 6 | 476 | 52,748.79 | 9.13 | 48.41 | 71.89 |
| Gh_A12G0114 | GhRLCK57A | 2037 | 1200 | 6 | 399 | 43,784.99 | 9.15 | 36.77 | 78.95 |
| Gh_A12G1525 | GhRLCK69A | 3644 | 1266 | 6 | 421 | 46,018.49 | 9.73 | 41.96 | 77.41 |
| Gh_A12G1932 | GhRLCK24A | 2259 | 1170 | 5 | 389 | 43,452.70 | 9.67 | 38.02 | 81.18 |
| Gh_A13G0455 | GhRLCK38A | 3979 | 1461 | 6 | 486 | 54,009.00 | 9.09 | 39.93 | 71.46 |
| Gh_A13G0572 | GhRLCK56A | 2997 | 1233 | 6 | 410 | 45,260.38 | 9.69 | 36.86 | 77.80 |
| Gh_A13G0635 | GhRLCK70A | 2995 | 1263 | 6 | 420 | 45,829.34 | 9.54 | 39.99 | 79.19 |
| Gh_A13G0755 | GhRLCK18A | 2701 | 1281 | 5 | 426 | 47,179.57 | 6.30 | 42.63 | 79.27 |
| Gh_A13G1685 | GhRLCK52A | 1680 | 987 | 5 | 328 | 36,794.49 | 6.63 | 44.02 | 71.98 |
| Gh_A13G1811 | GhRLCK43A | 5138 | 1323 | 6 | 440 | 49,149.66 | 6.58 | 36.73 | 72.73 |
| Gh_A13G2001 | GhRLCK15A | 2461 | 1260 | 5 | 419 | 46,393.73 | 8.02 | 24.60 | 77.28 |
| Gh_D01G0203 | GhRLCK13D | 4288 | 1392 | 5 | 463 | 50,970.19 | 6.52 | 35.26 | 71.86 |
| Gh_D01G0204 | GhRLCK12D | 4545 | 1392 | 5 | 463 | 51,216.57 | 7.63 | 35.17 | 73.11 |
| Gh_D01G0620 | GhRLCK1D | 1877 | 1161 | 5 | 386 | 42,940.31 | 4.99 | 40.17 | 74.82 |
| Gh_D01G0869 | GhRLCK22D | 3258 | 1146 | 5 | 381 | 42,271.09 | 8.44 | 37.20 | 82.91 |
| Gh_D01G1270 | GhRLCK35D | 1882 | 1077 | 7 | 358 | 40,378.33 | 9.12 | 39.34 | 83.83 |
| Gh_D01G2115 | GhRLCK11D | 3739 | 1200 | 5 | 399 | 44,211.89 | 9.48 | 32.54 | 78.02 |
| Gh_D01G2170 | GhRLCK63D | 2700 | 1116 | 6 | 372 | 41,389.03 | 9.54 | 41.45 | 79.65 |
| Gh_D01G2270 | GhRLCK8D | 3044 | 1554 | 5 | 517 | 56,787.69 | 9.02 | 37.16 | 66.63 |
| Gh_D02G0019 | GhRLCK45D | 3446 | 1281 | 6 | 426 | 47,210.73 | 8.91 | 35.55 | 82.39 |
| Gh_D02G1680 | GhRLCK36D | 2136 | 1128 | 7 | 375 | 42,423.62 | 8.89 | 45.91 | 85.52 |
| Gh_D02G1980 | GhRLCK55D | 2018 | 1233 | 6 | 410 | 45,604.95 | 9.53 | 34.19 | 76.80 |
| Gh_D03G0018 | GhRLCK58D | 2263 | 1251 | 6 | 416 | 46,384.36 | 8.81 | 33.93 | 74.04 |
| Gh_D03G0702 | GhRLCK68D | 1942 | 1434 | 6 | 477 | 51,918.05 | 8.06 | 52.13 | 89.08 |
| Gh_D03G1405 | GhRLCK46D | 7154 | 1200 | 7 | 399 | 44,896.47 | 6.61 | 37.33 | 88.72 |
| Gh_D03G1604 | GhRLCK47D | 2179 | 1338 | 5 | 445 | 49,394.48 | 9.61 | 36.70 | 81.30 |
| Gh_D04G0075 | GhRLCK43D | 4213 | 1185 | 5 | 394 | 44,283.03 | 6.35 | 35.79 | 75.25 |
| Gh_D04G0266 | GhRLCK19D | 1221 | 1128 | 2 | 375 | 42,145.62 | 8.20 | 63.01 | 60.08 |
| Gh_D04G0458 | GhRLCK40D | 3425 | 1479 | 6 | 492 | 54,423.71 | 9.35 | 42.40 | 69.59 |
| Gh_D04G1073 | GhRLCK32D | 3150 | 1368 | 5 | 455 | 51,269.75 | 9.64 | 41.39 | 79.89 |
| Gh_D04G1772 | GhRLCK34D | 3351 | 1080 | 7 | 359 | 40,453.42 | 9.05 | 42.48 | 83.62 |
| Gh_D05G0134 | GhRLCK41D | 1700 | 1290 | 6 | 369 | 41,304.47 | 9.59 | 36.56 | 88.48 |
| Gh_D05G0345 | GhRLCK53D | 1704 | 1356 | 5 | 451 | 51,241.70 | 9.07 | 40.22 | 78.47 |
| Gh_D05G0730 | GhRLCK66D | 2424 | 1149 | 6 | 382 | 41,999.77 | 9.45 | 28.69 | 78.93 |
| Gh_D05G0928 | GhRLCK28D | 1396 | 990 | 4 | 329 | 37,551.18 | 8.28 | 34.95 | 93.62 |
| Gh_D05G0929 | GhRLCK60D | 2365 | 1218 | 6 | 405 | 44,664.62 | 9.53 | 37.32 | 75.46 |
| Gh_D05G1341 | GhRLCK48D | 1728 | 1344 | 5 | 447 | 50,614.22 | 3.39 | 44.57 | 87.20 |
| Gh_D05G2814 | GhRLCK30D | 2122 | 1284 | 4 | 427 | 48,170.15 | 9.55 | 44.13 | 83.77 |
| Gh_D05G2857 | GhRLCK10D | 3012 | 1152 | 5 | 383 | 42,268.30 | 9.20 | 27.68 | 81.78 |
| Gh_D06G0789 | GhRLCK54D | 1610 | 1332 | 4 | 443 | 49,756.95 | 9.05 | 42.82 | 79.46 |
| Gh_D06G0842 | GhRLCK50D | 1887 | 1296 | 5 | 431 | 48,285.41 | 7.57 | 30.46 | 89.33 |
| Gh_D06G1617 | GhRLCK4D | 1617 | 1167 | 5 | 388 | 43,262.31 | 5.72 | 37.23 | 86.21 |
| Gh_D07G0159 | GhRLCK51D | 1791 | 1347 | 4 | 448 | 50,522.14 | 8.55 | 33.40 | 88.75 |
| Gh_D07G0376 | GhRLCK42D | 3004 | 1284 | 6 | 427 | 47,648.11 | 9.29 | 33.12 | 78.99 |
| Gh_D08G0016 | GhRLCK14D | 1854 | 1269 | 5 | 422 | 46,941.23 | 8.36 | 34.34 | 75.12 |
| Gh_D08G1756 | GhRLCK23D | 1869 | 1170 | 4 | 389 | 43,814.90 | 9.49 | 43.00 | 80.15 |
| Gh_D09G0202 | GhRLCK37D | 3015 | 1479 | 5 | 480 | 53,840.51 | 9.54 | 46.34 | 75.38 |
| Gh_D09G0393 | GhRLCK21D | 3290 | 1284 | 6 | 427 | 47,548.21 | 8.25 | 25.63 | 78.13 |
| Gh_D09G0598 | GhRLCK6D | 5006 | 1545 | 5 | 514 | 56,753.74 | 9.20 | 34.63 | 67.20 |
| Gh_D09G1021 | GhRLCK67D | 3802 | 1236 | 6 | 411 | 45,326.54 | 9.30 | 26.71 | 76.42 |
| Gh_D09G1396 | GhRLCK33D | 2382 | 1299 | 4 | 432 | 49,364.69 | 9.52 | 41.66 | 80.97 |
| Gh_D09G2328 | GhRLCK5D | 2534 | 1506 | 5 | 501 | 55,342.20 | 8.90 | 37.84 | 70.70 |
| Gh_D10G0142 | GhRLCK3D | 3818 | 1209 | 6 | 402 | 44,636.94 | 6.77 | 27.78 | 93.38 |
| Gh_D10G0523 | GhRLCK49D | 1677 | 1368 | 5 | 455 | 51,016.70 | 9.17 | 35.66 | 90.86 |
| Gh_D10G1669 | GhRLCK7D | 4019 | 1548 | 5 | 515 | 56,700.66 | 9.07 | 37.10 | 68.21 |
| Gh_D10G2126 | GhRLCK9D | 2466 | 1152 | 5 | 383 | 42,414.34 | 8.96 | 28.25 | 80.26 |
| Gh_D10G2531 | GhRLCK31D | 2505 | 1290 | 4 | 429 | 48,987.01 | 9.71 | 42.53 | 82.73 |
| Gh_D11G0278 | GhRLCK25D | 1816 | 1164 | 4 | 387 | 43,406.49 | 9.65 | 34.94 | 79.33 |
| Gh_D11G0666 | GhRLCK20D | 2669 | 1242 | 5 | 413 | 45,792.30 | 8.96 | 32.93 | 74.41 |
| Gh_D11G0732 | GhRLCK38D | 3096 | 1431 | 6 | 476 | 52,729.83 | 9.26 | 47.90 | 71.28 |
| Gh_D11G0788 | GhRLCK27D | 2199 | 1317 | 4 | 438 | 49,406.51 | 9.37 | 40.98 | 78.84 |
| Gh_D11G1413 | GhRLCK16D | 1667 | 1290 | 5 | 429 | 47,135.87 | 5.81 | 36.45 | 74.66 |
| Gh_D11G2878 | GhRLCK2D | 1816 | 1134 | 5 | 377 | 42,497.91 | 8.43 | 39.47 | 92.28 |
| Gh_D11G3068 | GhRLCK17D | 3797 | 1677 | 5 | 558 | 60,534.28 | 6.85 | 45.87 | 63.51 |
| Gh_D12G0126 | GhRLCK57D | 2040 | 1200 | 6 | 399 | 43,852.03 | 9.15 | 38.26 | 79.67 |
| Gh_D12G1647 | GhRLCK69D | 3772 | 1266 | 6 | 421 | 46,004.51 | 9.76 | 42.23 | 76.72 |
| Gh_D12G2113 | GhRLCK24D | 1859 | 1161 | 4 | 386 | 43,117.45 | 9.73 | 40.05 | 81.32 |
| Gh_D13G0556 | GhRLCK56D | 2958 | 1233 | 6 | 410 | 45,274.36 | 9.56 | 36.25 | 79.00 |
| Gh_D13G0697 | GhRLCK39D | 3979 | 1461 | 6 | 486 | 53,902.97 | 9.20 | 40.46 | 71.67 |
| Gh_D13G0752 | GhRLCK70D | 2944 | 1263 | 6 | 420 | 45,827.33 | 9.54 | 40.94 | 78.50 |
| Gh_D13G0942 | GhRLCK18D | 2743 | 1281 | 5 | 426 | 47,120.45 | 6.00 | 42.66 | 78.83 |
| Gh_D13G2164 | GhRLCK44D | 5149 | 1281 | 6 | 426 | 47,696.18 | 8.40 | 37.30 | 73.97 |
| Gh_D13G2375 | GhRLCK29D | 2178 | 1422 | 4 | 473 | 53,422.69 | 8.06 | 34.63 | 81.59 |
| Gh_D13G2376 | GhRLCK61D | 2179 | 1209 | 6 | 402 | 44,454.33 | 9.43 | 33.59 | 76.89 |
| Gh_D13G2400 | GhRLCK15D | 2466 | 1260 | 5 | 419 | 46,465.84 | 8.48 | 23.98 | 77.04 |
| Gh_D13G2490 | GhRLCK52D | 1737 | 1383 | 5 | 460 | 51,581.21 | 6.66 | 46.09 | 74.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cen, Y.; Geng, S.; Gao, L.; Wang, X.; Yan, X.; Hou, Y.; Wang, P. Genome-Wide Identification and Expression Analysis of RLCK-VII Subfamily Genes Reveal Their Roles in Stress Responses of Upland Cotton. Plants 2023, 12, 3170. https://doi.org/10.3390/plants12173170
Cen Y, Geng S, Gao L, Wang X, Yan X, Hou Y, Wang P. Genome-Wide Identification and Expression Analysis of RLCK-VII Subfamily Genes Reveal Their Roles in Stress Responses of Upland Cotton. Plants. 2023; 12(17):3170. https://doi.org/10.3390/plants12173170
Chicago/Turabian StyleCen, Yuhan, Shiyi Geng, Linying Gao, Xinyue Wang, Xin Yan, Yuxia Hou, and Ping Wang. 2023. "Genome-Wide Identification and Expression Analysis of RLCK-VII Subfamily Genes Reveal Their Roles in Stress Responses of Upland Cotton" Plants 12, no. 17: 3170. https://doi.org/10.3390/plants12173170
APA StyleCen, Y., Geng, S., Gao, L., Wang, X., Yan, X., Hou, Y., & Wang, P. (2023). Genome-Wide Identification and Expression Analysis of RLCK-VII Subfamily Genes Reveal Their Roles in Stress Responses of Upland Cotton. Plants, 12(17), 3170. https://doi.org/10.3390/plants12173170





