Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation
Abstract
:1. Introduction
2. Results
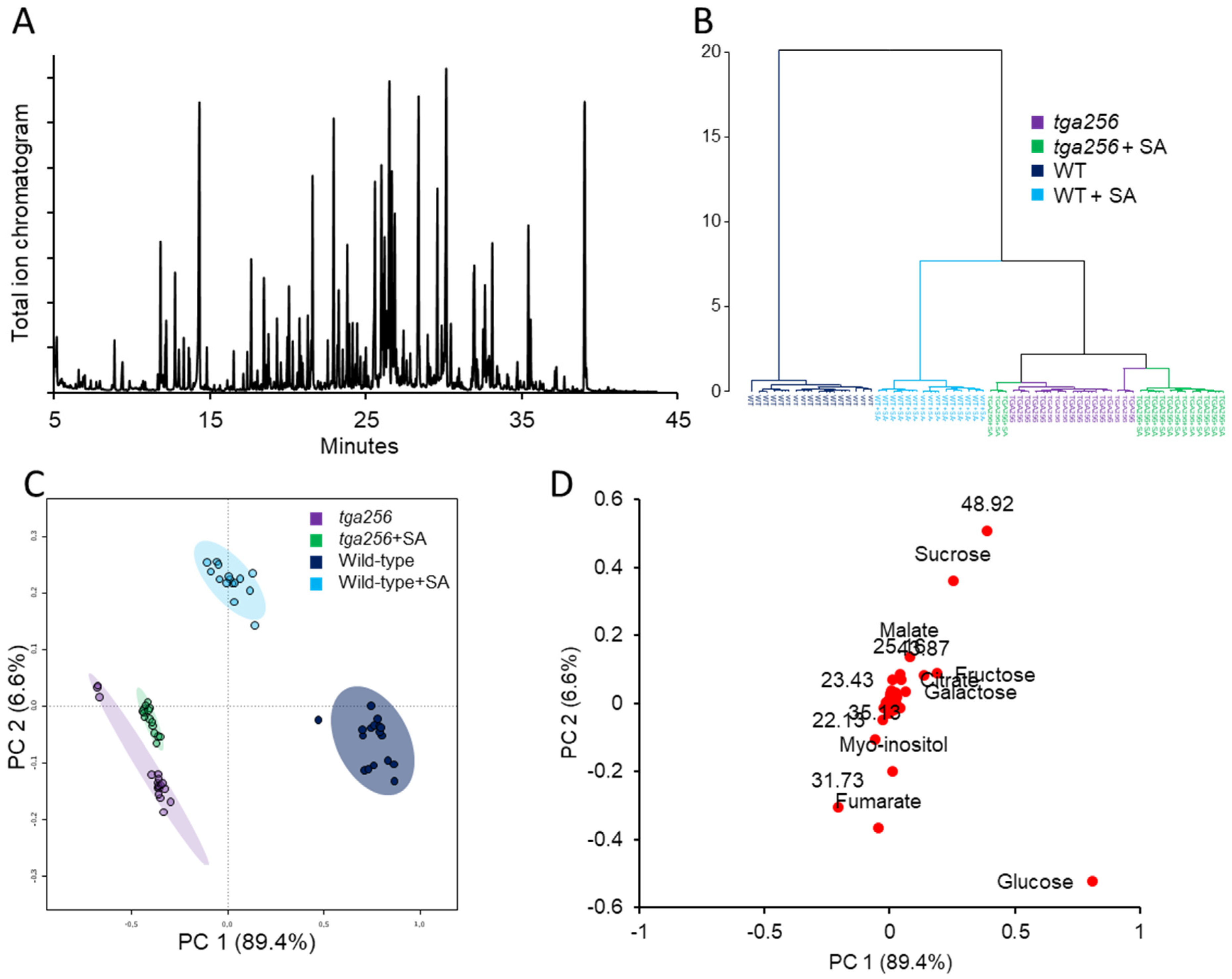
2.1. Clade II TGAs Maintain Sugar Homeostasis by Buffering SA-Induced Suppression of Central Metabolism
2.2. The MEP Pathway Is Not under Negative Feedback Regulation by SA
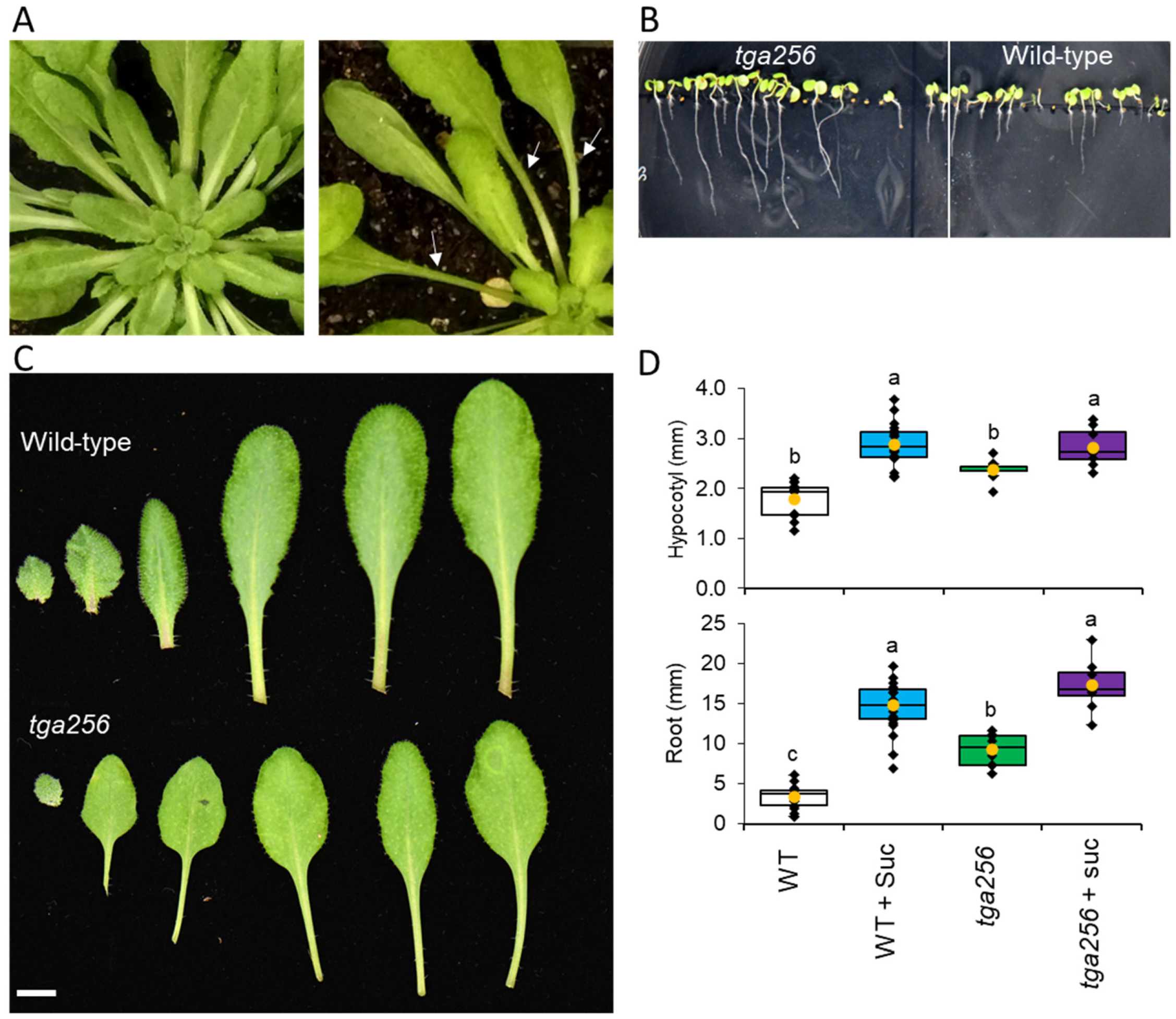
2.3. The tga256 Mutant Displays Elongated Petioles, Hypocotyls, and Roots
2.4. The tga256 Triple Mutant Is Deficient in Gibberellin but Has Normal Auxin Levels
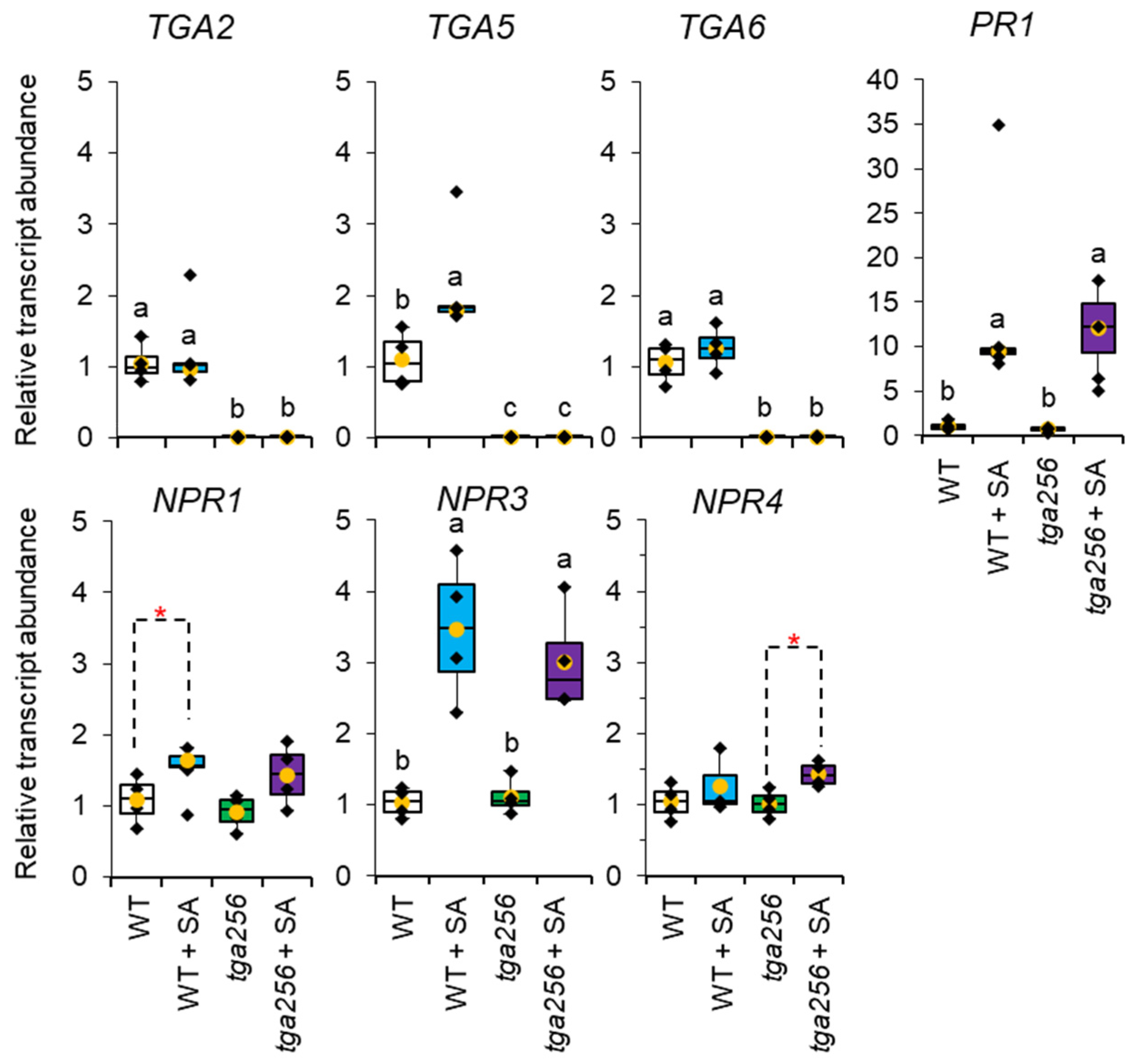
2.5. NPR3, NPR4, and PR1 Transcripts Are Responsive to SA Induction Independently of TGA256
3. Discussion
3.1. TGA2/5/6 Negative Regulatory Functions Are Required for Sugar Homeostasis
3.2. Altered Growth Patterns in tga256 Result from a Disruption in Sucrose Supply
3.3. Role of Small Metabolite Signals in Clade II TGA Defense Signaling
4. Materials and Methods
4.1. Plant Growth and Treatments
4.2. Measurement of Hypocotyl Length
4.3. GCMS Analysis
4.4. LC-MS/MS Analysis
4.5. Transcript Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Després, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tessaro, M.J.; Lassner, M.; Li, X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 2003, 15, 2647–2653. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467.e1415. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.; Urzúa, T.; Jelenska, J.; Sbarbaro, C.; Seguel, A.; Duarte, Y.; Greenberg, J.T.; Holuigue, L.; Blanco-Herrera, F.; Herrera-Vásquez, A. The TGA transcription factors from clade II negatively regulate the salicylic acid accumulation in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 11631. [Google Scholar] [CrossRef] [PubMed]
- Rivasseau, C.; Seemann, M.; Boisson, A.-M.; Streb, P.; Gout, E.; Douce, R.; Rohmer, M.; Bligny, R. Accumulation of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate in illuminated plant leaves at supraoptimal temperatures reveals a bottleneck of the prokaryotic methylerythritol 4-phosphate pathway of isoprenoid biosynthesis. Plant Cell Environ. 2009, 32, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Estrada-Tejedor, R.; Volke, D.C.; Phillips, M.A.; Gershenzon, J.; Wright, L.P. Negative regulation of plastidial isoprenoid pathway by herbivore-induced β-cyclocitral in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2008747118. [Google Scholar] [CrossRef]
- Gil, M.J.; Coego, A.; Mauch-Mani, B.; Jorda, L.; Vera, P. The Arabidopsis csb3 mutant reveals a regulatory link between salicylic acid-mediated disease resistance and the methyl-erythritol 4-phosphate pathway. Plant J. 2005, 44, 155–166. [Google Scholar] [CrossRef]
- Hussain, R.M.; Sheikh, A.H.; Haider, I.; Quareshy, M.; Linthorst, H.J. Arabidopsis WRKY50 and TGA transcription factors synergistically activate expression of PR1. Front. Plant Sci. 2018, 9, 930. [Google Scholar] [CrossRef]
- Herrera-Vásquez, A.; Fonseca, A.; Ugalde, J.M.; Lamig, L.; Seguel, A.; Moyano, T.C.; Gutiérrez, R.A.; Salinas, P.; Vidal, E.A.; Holuigue, L. TGA class II transcription factors are essential to restrict oxidative stress in response to UV-B stress in Arabidopsis. J. Exp. Bot. 2020, 72, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, T.; Sun, Y.; Zhang, Y.; Radojičić, A.; Ding, Y.; Tian, H.; Huang, X.; Lan, J.; Chen, S.; et al. Diverse Roles of the Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Plant Immunity. Plant Cell 2020, 32, 4002–4016. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; La Camera, S.; Lamotte, O.; Métraux, J.-P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef]
- Kachroo, P.; Burch-Smith, T.M.; Grant, M. An Emerging Role for Chloroplasts in Disease and Defense. Annu. Rev. Phytopathol. 2021, 59, 423–445. [Google Scholar] [CrossRef]
- Lemos, M.; Xiao, Y.; Bjornson, M.; Wang, J.-z.; Hicks, D.; Souza, A.d.; Wang, C.-Q.; Yang, P.; Ma, S.; Dinesh-Kumar, S.; et al. The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J. Exp. Bot. 2016, 67, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Hu, Y.; Liu, H.; He, M.; Yang, Z.; Kong, F.; Liu, X.; Hou, X. DELLA and EDS1 form a feedback regulatory module to fine-tune plant growth–defense tradeoff in Arabidopsis. Mol. Plant. 2019, 12, 1485–1498. [Google Scholar] [CrossRef]
- Phillips, M.A.; León, P.; Boronat, A.; Rodríguez-Concepción, M. The plastidial MEP pathway: Unified nomenclature and resources. Trends Plant Sci. 2008, 13, 619–623. [Google Scholar] [CrossRef]
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.K.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- González-Cabanelas, D.; Wright, L.P.; Paetz, C.; Onkokesung, N.; Gershenzon, J.; Rodríguez Concepción, M.; Phillips, M.A. The diversion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate from the 2-C-methyl-D-erythritol 4-phosphate pathway to hemiterpene glycosides mediates stress responses in Arabidopsis thaliana. Plant J. 2015, 82, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.; Xiao, Y.; Wang, J.-Z.; Baidoo, E.E.; Keasling, J.D.; Shen, Z.; Briggs, S.P.; Dehesh, K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 6212–6217. [Google Scholar] [CrossRef] [PubMed]
- Benn, G.; Bjornson, M.; Ke, H.; De Souza, A.; Balmond, E.I.; Shaw, J.T.; Dehesh, K. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. USA 2016, 113, 8855–8860. [Google Scholar] [CrossRef]
- Jiang, J.; Rodriguez-Furlan, C.; Wang, J.-Z.; de Souza, A.; Ke, H.; Pasternak, T.; Lasok, H.; Ditengou, F.A.; Palme, K.; Dehesh, K. Interplay of the two ancient metabolites auxin and MEcPP regulates adaptive growth. Nat. Commun. 2018, 9, 2262. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Primavesi, L.F.; Jhurreea, D.; Zhang, Y. Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 2008, 59, 417–441. [Google Scholar] [CrossRef]
- Zander, M.; Thurow, C.; Gatz, C. TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 2014, 165, 1671–1683. [Google Scholar] [CrossRef]
- Alabadí, D.; Gallego-Bartolomé, J.; Orlando, L.; García-Cárcel, L.; Rubio, V.; Martínez, C.; Frigerio, M.; Iglesias-Pedraz, J.M.; Espinosa, A.; Deng, X.W.; et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008, 53, 324–335. [Google Scholar] [CrossRef]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 2008, 132, 417–425. [Google Scholar] [CrossRef]
- Chen, J.; Mohan, R.; Zhang, Y.; Li, M.; Chen, H.; Palmer, I.A.; Chang, M.; Qi, G.; Spoel, S.H.; Mengiste, T.; et al. NPR1 promotes its own and target gene expression in plant defense by recruiting CDK81. Plant Physiol. 2019, 181, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; Chen, S.; Imkampe, J.; Thurow, C.; Gatz, C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL Motif. Mol. Plant. 2012, 5, 831–840. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, J.M.; Lamig, L.; Herrera-Vásquez, A.; Fuchs, P.; Homagk, M.; Kopriva, S.; Müller-Schüssele, S.J.; Holuigue, L.; Meyer, A.J. A dual role for glutathione transferase U7 in plant growth and protection from methyl viologen-induced oxidative stress. Plant Physiol. 2021, 187, 2451–2468. [Google Scholar] [CrossRef]
- Kuai, X.; MacLeod, B.J.; Després, C. Integrating data on the Arabidopsis NPR1/NPR3/NPR4 salicylic acid receptors; a differentiating argument. Front. Plant Sci. 2015, 6, 235. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006, 48, 647–656. [Google Scholar] [CrossRef]
- Innes, R. The positives and negatives of NPR: A unifying model for salicylic acid signaling in plants. Cell 2018, 173, 1314–1315. [Google Scholar] [CrossRef]
- Schweiger, R.; Heise, A.-M.; Persicke, M.; Müller, C. Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant Cell Environ. 2014, 37, 1574–1585. [Google Scholar] [CrossRef]
- Wei, X.; Vrieling, K.; Kim, H.K.; Mulder, P.P.J.; Klinkhamer, P.G.L. Application of methyl jasmonate and salicylic acid lead to contrasting effects on the plant’s metabolome and herbivory. Plant Science 2021, 303, 110784. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, Y.; Yang, C.; Shi, H. The combination of transcriptome and metabolome reveals the molecular mechanism by which topping and salicylic acid treatment affect the synthesis of alkaloids in Nicotiana tabacum L. All Life 2022, 15, 147–159. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant. 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Milmanda, G.L.; Ballaré, C.L. Shade avoidance: Expanding the color and hormone palette. Trends Plant Sci. 2021, 26, 509–523. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Sellaro, R.; Zurbriggen, M.D.; Casal, J.J. Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 2017, 69, 213–228. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.-L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- de Lucas, M.; Davière, J.-M.; Rodríguez-Falcón, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blázquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef]
- Ciereszko, I. Regulatory roles of sugars in plant growth and development. Acta Soc. Bot. Pol. 2018, 87, 3583. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.S.; Sharma, M.; Laxmi, A. Role of sugar and auxin crosstalk in plant growth and development. Physiol. Plant. 2022, 174, e13546. [Google Scholar] [CrossRef]
- Li, L.; Sheen, J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016, 33, 116–125. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dédaldéchamp, F.; Perez-Garcia, M.-D.; Ogé, L.; Hamama, L.; Atanassova, R. The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, P.; Jha, G. Alterations in plant sugar metabolism: Signatory of pathogen attack. Planta 2019, 249, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Bezrutczyk, M.; Yang, J.; Eom, J.-S.; Prior, M.; Sosso, D.; Hartwig, T.; Szurek, B.; Oliva, R.; Vera-Cruz, C.; White, F.F.; et al. Sugar flux and signaling in plant–microbe interactions. Plant J. 2018, 93, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant SWEETs: From sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gautam, T. SWEET genes and TAL effectors for disease resistance in plants: Present status and future prospects. Mol. Plant Pathol. 2021, 22, 1014–1026. [Google Scholar] [CrossRef]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.-H.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector–mediated induction of a SWEET sugar transporter in Cassava. Mol. Plant Microbe Interact. 2014, 27, 1186–1198. [Google Scholar] [CrossRef]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.-P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef]
- Fichtner, F.; Lunn, J.E. The Role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.G.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 2017, 92, 611–623. [Google Scholar] [CrossRef]
- Schluepmann, H.; Pellny, T.; van Dijken, A.; Smeekens, S.; Paul, M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6849–6854. [Google Scholar] [CrossRef] [PubMed]
- Schluepmann, H.; van Dijken, A.; Aghdasi, M.; Wobbes, B.; Paul, M.; Smeekens, S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 2004, 135, 879–890. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, X.; Yang, X.; Zhao, T.; An, X.; Chen, Z. Characterization and expression pattern of the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase gene families in Populus. Int. J. Biol. Macromol. 2021, 187, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Dan, Y.; Niu, Y.; Wang, C.; Yan, M.; Liao, W. Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase (TPS) gene family in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e11398. [Google Scholar] [CrossRef]
- Liu, K.; Zhou, Y. Genome-wide identification of the trehalose-6-phosphate synthase gene family in sweet orange (Citrus sinensis) and expression analysis in response to phytohormones and abiotic stresses. PeerJ 2022, 10, e13934. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.E.; Duggan, P.; Bergman, M.E.; Cobo-López, D.; Davis, B.; Bajwa, I.; Phillips, M.A. Design and fabrication of an improved dynamic flow cuvette for 13CO2 labeling in Arabidopsis plants. Plant Methods 2022, 18, 1–13. [Google Scholar] [CrossRef]
- Bergman, M.E.; Evans, S.E.; Davis, B.; Hamid, R.; Bajwa, I.; Jayathilake, A.; Chahal, A.K.; Phillips, M.A. An Arabidopsis GCMS chemical ionization technique to quantify adaptive responses in central metabolism. Plant Physiol. 2022, 189, 2072–2090. [Google Scholar] [CrossRef]
- Hummel, J.; Strehmel, N.; Selbig, J.; Walther, D.; Kopka, J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 2010, 6, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; D’Auria, J.C.; Luck, K.; Gershenzon, J. Evaluation of Candidate Reference Genes for Real-Time Quantitative PCR of Plant Samples Using Purified cDNA as Template. Plant Mol. Biol. Rep. 2009, 27, 407–416. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergman, M.E.; Evans, S.E.; Kuai, X.; Franks, A.E.; Despres, C.; Phillips, M.A. Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation. Plants 2023, 12, 3284. https://doi.org/10.3390/plants12183284
Bergman ME, Evans SE, Kuai X, Franks AE, Despres C, Phillips MA. Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation. Plants. 2023; 12(18):3284. https://doi.org/10.3390/plants12183284
Chicago/Turabian StyleBergman, Matthew E., Sonia E. Evans, Xiahezi Kuai, Anya E. Franks, Charles Despres, and Michael A. Phillips. 2023. "Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation" Plants 12, no. 18: 3284. https://doi.org/10.3390/plants12183284
APA StyleBergman, M. E., Evans, S. E., Kuai, X., Franks, A. E., Despres, C., & Phillips, M. A. (2023). Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation. Plants, 12(18), 3284. https://doi.org/10.3390/plants12183284





