Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks
Abstract
:1. Introduction
2. Results
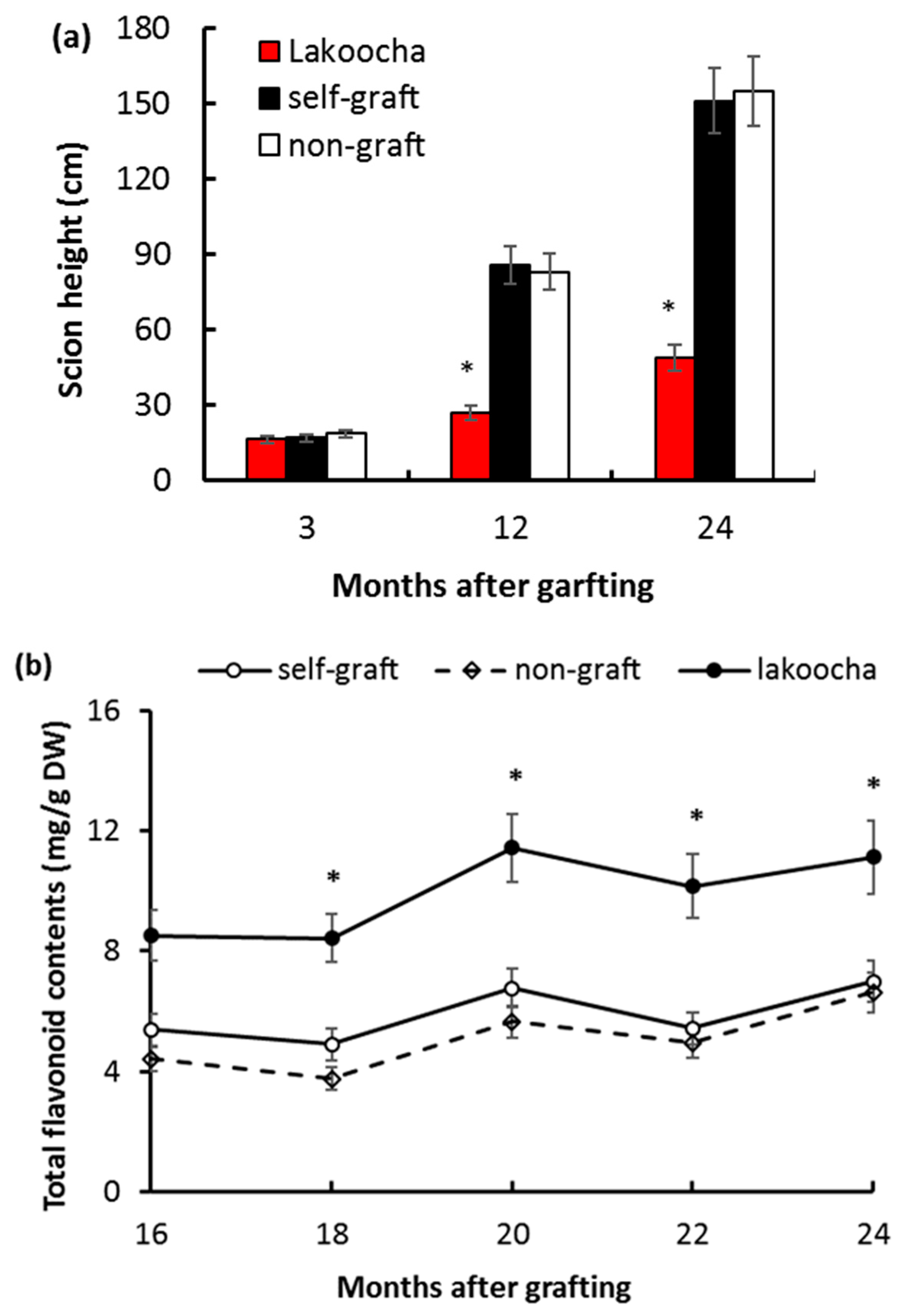
2.1. Effects of Rootstocks on Height and Total Flavonoid Content in Breadfruit Scions
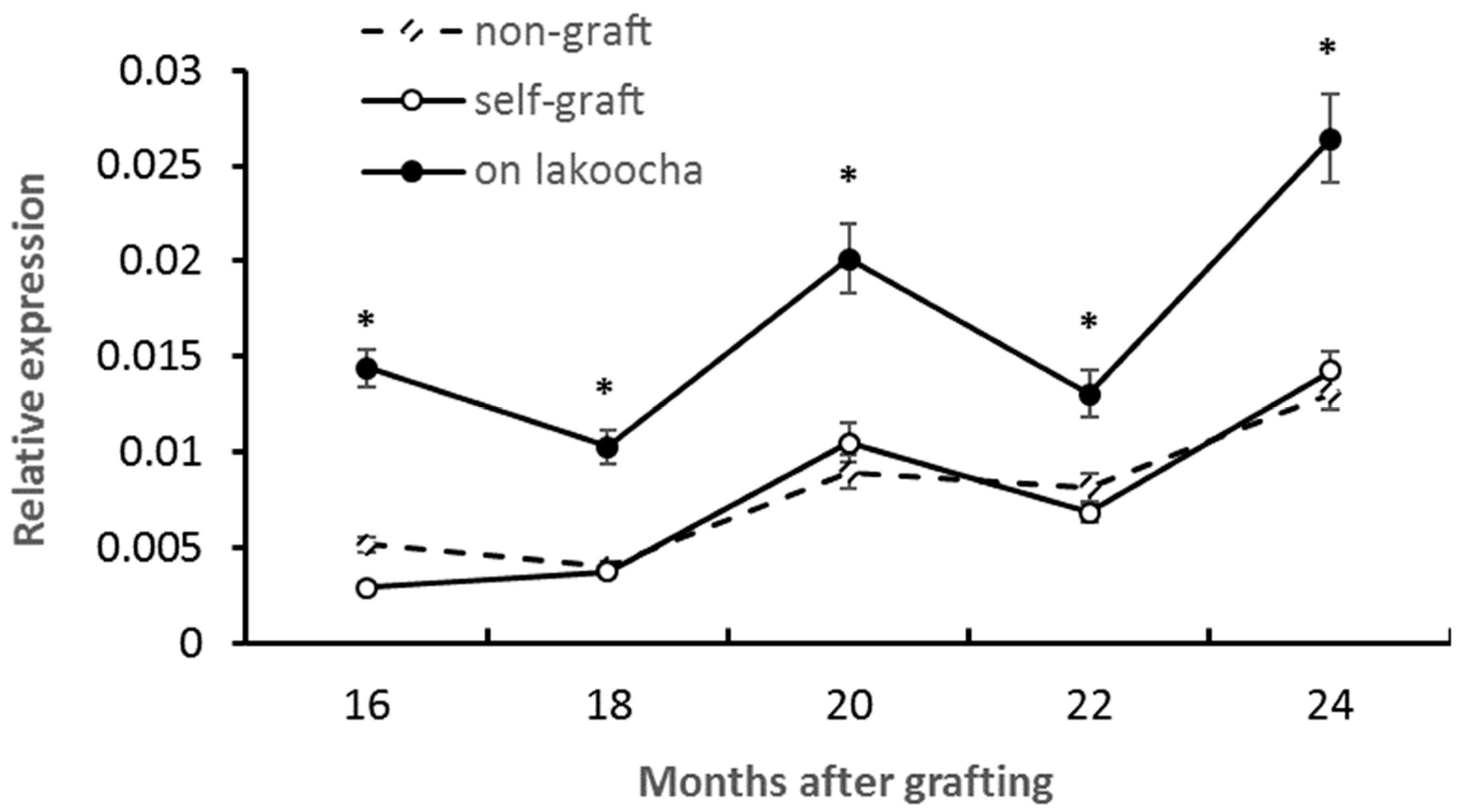
2.2. Isolation and Expression Analysis of the Chalcone Synthase Gene in Breadfruit Scions
2.3. Isolation and Expression Analysis of the Dihydroflavonol 4-Reductase Gene in Breadfruit Scions
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Determination of Total Flavonoid Content
4.3. Cloning Chalcone Synthase and Dihydroflavonol 4-Reductase cDNA
4.4. Quantitative Real-Time PCR
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, A.M.P.; Ragone, D.; Tavana, N.G.; Bernotas, D.W.; Murch, S.J. Beyond the bounty: Breadfruit (Artocarpus altilis) for food security and novel foods in the 21st century. Ethnobot. Res. Appl. 2011, 9, 129–149. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Characterisation of breadfruit (Artocarpus altilis) plants growing on Lakoocha (A. lakoocha) rootstocks. Horticulturae 2022, 8, 916. [Google Scholar] [CrossRef]
- Gregory, P.; Atkinson, C.; Bengough, A.; Else, M.; Fernández-Fernández, F.; Harrison, R.; Schmidt, S. Contributions of roots and rootstocks to sustainable, intensified crop production. J. Exp. Bot. 2013, 64, 1209–1222. [Google Scholar] [CrossRef]
- Hayat, F.; Li, J.; Iqbal, S.; Khan, U.; Ali, N.; Peng, Y.; Hong, L.; Asghar, S.; Javed, H.; Li, C.; et al. Hormonal interactions underlying rootstock-induced vigor control in horticultural crops. Appl. Sci. 2023, 13, 1237. [Google Scholar] [CrossRef]
- Else, M.A.; Taylor, J.M.; Young, S.; Atkinson, C.J. The effect of the graft union on hormonal and ionic signalling between rootstocks and scions of grafted apple (Malus pumila L. Mill.). Environ. Exp. Bot. 2018, 156, 325–336. [Google Scholar] [CrossRef]
- Lochard, R.G.; Schneider, G. Stock and scion growth relationships and the dwarfing mechanism in apple. Hortic. Rev. 2011, 3, 315–375. [Google Scholar] [CrossRef]
- Song, C.; Zhang, D.; Zhang, J.; Zheng, L.; Zhao, C.; Ma, J.; An, N.; Han, M. Expression analysis of key auxin synthesis, transport, and metabolism genes in different young dwarfing apple trees. Acta Physiol. Plant. 2016, 38, 43. [Google Scholar] [CrossRef]
- Chen, Y.; An, X.; Zhao, D.; Li, E.; Ma, R.; Li, Z.; Cheng, C. Transcription profiles reveal sugar and hormone signaling pathways mediating tree branch architecture in apple (Malus domestica Borkh.) grafted on different rootstocks. PLoS ONE 2020, 15, e0236530. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Zhuang, W.B.; Tu, X.T.; Gao, Z.H.; Xiong, A.S.; Yu, X.Y.; Li, X.H.; Li, F.H.; Qu, S.C. Transcriptomic analysis of interstock-induced dwarfism in Sweet Persimmon (Diospyros kaki Thunb.). Hortic. Res. 2019, 6, 51. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Differential transcription pathways associated with rootstock-induced dwarfing in breadfruit (Artocarpus altilis) scions. BMC Plant Biol. 2021, 21, 261. [Google Scholar] [CrossRef]
- Cong, L.; Ling, H.; Liu, S.; Wang, A.; Zhai, R.; Yang, C.; Wang, Z.; Xu, L. ‘Yunnan’ quince rootstock promoted flower bud formation of ‘Abbé Fetel’ pear by altering hormone levels and PbAGL9 expression. J. Plant Physiol. 2023, 282, 153924. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Li, J.; Liu, M.M.; Yao, Q.; Chen, J.Z. Transcriptome profiling to understand the effect of citrus rootstocks on the growth of ‘Shatangju’ mandarin. PLoS ONE 2017, 12, e0169897. [Google Scholar] [CrossRef] [PubMed]
- Van Hooijdonk, B.M.; Woolley, D.J.; Warrington, I.J.; Tustin, D.S. Initial alteration of scion architecture by dwarfing apple rootstocks may involve shoot-root-shoot signalling by auxin, gibberellin, and cytokinin. J. Hortic. Sci. Biotech. 2010, 85, 59–65. [Google Scholar] [CrossRef]
- Yin, R.H.; Han, K.; Heller, W.; Albert, A.; Dobrev, P.I.; Zazimalova, E.; Schaffner, A.R. Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 2014, 201, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.M.; McAtee, P.A.; Waite, C.N.; Boldingh, H.L.; McGhie, T.K. Apple dwarfing rootstocks exhibit an imbalance in carbohydrate allocation and reduced cell growth and metabolism. Hortic. Res. 2017, 4, 17009. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Zhang, H.; Galarneau, E.R.A.; Yang, Y.; Li, D.; Song, J.; Ma, C.; Zhang, S.; Wang, R. Metabolome analysis reveals important compounds related to dwarfing effect of interstock on scion in pear. Ann. Appl. Biol. 2021, 179, 108–122. [Google Scholar] [CrossRef]
- Hayat, F.; Li, J.; Liu, W.; Li, C.; Song, W.; Iqbal, S.; Khan, U.; Javed, H.U.; Altaf, M.A.; Tu, P.; et al. Influence of citrus rootstocks on scion growth, hormone levels, and metabolites profile of ‘Shatangju’ mandarin (Citrus reticulata Blanco). Horticulturae 2022, 8, 608. [Google Scholar] [CrossRef]
- Prassinos, C.; Ko, J.-H.; Lang, G.; Iezzoni, A.F.; Han, K.-H. Rootstock-induced dwarfing in cherries is caused by differential cessation of terminal meristem growth and is triggered by rootstock-specific gene regulation. Tree Physiol. 2009, 29, 927–936. [Google Scholar] [CrossRef]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin Transport and plant growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Lin, H.-X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Halbwirth, H. The creation and physiological relevance of divergent hydroxylation patterns in the flavonoid pathway. Int. J. Mol. Sci. 2010, 11, 595–621. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yu, Q.; Shen, W.; El Mohtar, C.A.; Zhao, X.; Gmitter, F.G. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Cheniany, M.; Ebrahimzadeh, H. Expression of chalcone synthase influences flavonoid content and frequency of rhizogenesis in microshoots of Juglans regia L. Plant Cell Tissue Organ Cult. (PCTOC) 2011, 109, 51–59. [Google Scholar] [CrossRef]
- El Euch, C.; Jay-Allemand, C.; Pastuglia, M.; Doumas, P.; Charpentier, J.P.; Capelli, P.; Jouanin, L. Expression of antisense chalcone synthase RNA in transgenic hybrid walnut microcuttings. Effect on flavonoid content and rooting ability. Plant Mol. Biol. 1998, 38, 467–479. [Google Scholar] [CrossRef]
- Chen, L.; Guo, H.; Lin, Y.; Cheng, H. Chalcone synthase EaCHS1 from Eupatorium adenophorum functions in salt stress tolerance in tobacco. Plant Cell Rep. 2015, 34, 885–894. [Google Scholar] [CrossRef]
- Shimada, N.; Sasaki, R.; Sato, S.; Kaneko, T.; Tabata, S.; Aoki, T.; Ayabe, S.-I. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J. Exp. Bot. 2005, 56, 2573–2585. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Puglisi, I.; Petrone, G. Gene characterization, analysis of expression and in vitro synthesis of dihydroflavonol 4-reductase from [Citrus sinensis (L.) Osbeck]. Phytochemistry 2006, 67, 684–695. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, Y.; Wang, H.; Tian, Z.; Xin, S.; Zhu, P. The dihydroflavonol 4-reductase BoDFR1 drives anthocyanin accumulation in pink-leaved ornamental kale. Theor. Appl. Genet. 2021, 134, 159–169. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Cai, X.; Shan, X.; Gao, R.; Yang, S.; Han, T.; Wang, S.; Wang, L.; Gao, X. Dihydroflavonol 4-reductase genes from Freesia hybrida play important and partially overlapping roles in the biosynthesis of flavonoids. Front. Plant Sci. 2017, 8, 428. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 2004, 134, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Gou, J.; Jia, Z.; Yang, L.; Sun, Y.; Xiao, X.; Song, F.; Luo, K. Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS ONE 2012, 7, e30364. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhou, N.; Wang, Y.; Sun, S.; Zhang, Y.; Ju, Z.; Yi, Y. Characterization and functional analysis of RdDFR1 regulation on flower color formation in Rhododendron delavayi. Plant Physiol. Biochem. 2021, 169, 203–210. [Google Scholar] [CrossRef]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Wang, C.; Zhi, S.; Liu, C.; Xu, F.; Zhao, A.; Wang, X.; Tang, X.; Li, Z.; Huang, P.; Yu, M. Isolation and characterization of a novel chalcone synthase gene family from mulberry. Plant Physiol. Biochem. 2017, 115, 107–118. [Google Scholar] [CrossRef]
- Goto-Yamamoto, N.; Wan, G.H.; Masaki, K.; Kobayashi, S. Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera). Plant Sci. 2002, 162, 867–872. [Google Scholar] [CrossRef]
- Yahyaa, M.; Ali, S.; Davidovich-Rikanati, R.; Ibdah, M.; Shachtier, A.; Eyal, Y.; Lewinsohn, E.; Ibdah, M. Characterization of three chalcone synthase-like genes from apple (Malus x domestica Borkh.). Phytochemistry 2017, 140, 125–133. [Google Scholar] [CrossRef]
- Anguraj Vadivel, A.K.; Krysiak, K.; Tian, G.; Dhaubhadel, S. Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L]Merr). BMC Plant Biol. 2018, 18, 325. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Yadav, S.K.; Ahuja, P.S. Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant Biotechnol. Rep. 2009, 3, 95–101. [Google Scholar] [CrossRef]
- Ferrer, J.-L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.Y.; Fukuma, K.; Kagami, J.; Yamazaki, Y.; Shibuya, M.; Ebizuka, Y.; Sankawa, U. Identification of amino acid residues important in the cyclization reactions of chalcone and stilbene synthases. Biochem. J. 2000, 350 Pt 1, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.T.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Buer, C.S.; Djordjevic, M.A. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Vassilevska-Ivanova, R.D.; Kraptchev, B.V.; Shtereva, L.A. An intergeneric sunflower line produced after cross Helianthus annuus x Echinacea purpurea. Genet. Resour. Crop Evol. 2015, 62, 829–836. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Yu, J.J.; Zhang, H.W.; Yang, Z.M. Relationship between the phenylpropanoid pathway and dwarfism of Paspalum seashore based on RNA-seq and iTRAQ. Int. J. Mol. Sci. 2021, 22, 9568. [Google Scholar] [CrossRef]
- Dare, A.P.; Tomes, S.; Jones, M.; McGhie, T.K.; Stevenson, D.E.; Johnson, R.A.; Greenwood, D.R.; Hellens, R.P. Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). Plant J. 2013, 74, 398–410. [Google Scholar] [CrossRef]
- Yin, Y.-C.; Hou, J.-M.; Tian, S.-K.; Yang, L.; Zhang, Z.-X.; Li, W.-D.; Liu, Y. Overexpressing chalcone synthase (CHS) gene enhanced flavonoids accumulation in Glycyrrhiza uralensis hairy roots. Bot. Lett. 2020, 167, 219–231. [Google Scholar] [CrossRef]
- Tsukaya, H.; Ohshima, T.; Naito, S.; Chino, M.; Komeda, Y. Sugar-dependent expression of the CHS-a gene for chalcone synthase from Petunia in transgenic Arabidopsis. Plant Physiol. 1991, 97, 1414–1421. [Google Scholar] [CrossRef]
- Li, Y.; Van Den Ende, W.; Rolland, F. Sucrose induction of anthocyanin biosynthesis is mediated by della. Mol. Plant 2014, 7, 570–572. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.; Li, H.; Fu, Y. Cooperative regulation of flavonoid and lignin biosynthesis in plants. Crit. Rev. Plant Sci. 2021, 40, 1–18. [Google Scholar] [CrossRef]
- Tworkoski, T.; Fazio, G. Effects of size-controlling apple rootstocks on growth, abscisic acid, and hydraulic conductivity of scion of different vigor. Int. J. Fruit Sci. 2015, 15, 369–381. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S.J.R. Expression of gibberellin metabolism genes and signalling components in dwarf phenotype of breadfruit (Artocarpus altilis) plants growing on marang (Artocarpus odoratissimus) rootstocks. Plants 2020, 9, 634. [Google Scholar] [CrossRef]
- Zhou, Y.; Underhill, S. Breadfruit (Artocarpus altilis) gibberellin 2-oxidase genes in stem elongation and abiotic stress response. Plant Physiol. Biochem. 2016, 98, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Underhill, S.J.R. Plasma membrane H-ATPase activity and graft success of breadfruit (Artocarpus altilis) onto interspecific rootstocks of marang (A. odoratissimus) and pedalai (A. sericicarpus). Plant Biol. 2018, 20, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Alirezalu, A.; Salehi, P.; Ahmadi, N.; Sonboli, A.; Aceto, S.; Hatami Maleki, H.; Ayyari, M. Flavonoids profile and antioxidant activity in flowers and leaves of hawthorn species (Crataegus spp.) from different regions of Iran. Int. J. Food Prop. 2018, 21, 452–470. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Underhill, S.J.R. Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks. Plants 2023, 12, 3285. https://doi.org/10.3390/plants12183285
Zhou Y, Underhill SJR. Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks. Plants. 2023; 12(18):3285. https://doi.org/10.3390/plants12183285
Chicago/Turabian StyleZhou, Yuchan, and Steven J. R. Underhill. 2023. "Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks" Plants 12, no. 18: 3285. https://doi.org/10.3390/plants12183285
APA StyleZhou, Y., & Underhill, S. J. R. (2023). Total Flavonoid Contents and the Expression of Flavonoid Biosynthetic Genes in Breadfruit (Artocarpus altilis) Scions Growing on Lakoocha (Artocarpus lakoocha) Rootstocks. Plants, 12(18), 3285. https://doi.org/10.3390/plants12183285





