Abstract
Vegetatively propagating aquatic angiosperms, the Lemnaceae family (duckweeds) represents valuable genetic resources for circular bioeconomics and other sustainable applications. Due to extremely fast growth and laborious cultivation of in vitro collections, duckweeds are an urgent subject for cryopreservation. We developed a robust and fast DMSO-free protocol for duckweed cryopreservation by vitrification. A single-use device was designed for sampling of duckweed fronds from donor culture, further spin-drying, and subsequent transferring to cryo-tubes with plant vitrification solution 3 (PVS3). Following cultivation in darkness and applying elevated temperatures during early regrowth stage, a specific pulsed illumination instead of a diurnal regime enabled successful regrowth after the cryopreservation of 21 accessions of Spirodela, Landoltia, Lemna, and Wolffia genera, including interspecific hybrids, auto- and allopolyploids. Genome size measurements revealed no quantitative genomic changes potentially caused by cryopreservation. The expression of CBF/DREB1 genes, considered as key factors in the development of freezing tolerance, was studied prior to cooling but was not linked with duckweed regrowth after rewarming. Despite preserving chlorophyll fluorescence after rewarming, the rewarmed fronds demonstrated nearly zero photosynthetic activity, which did not recover. The novel protocol provides the basis for future routine application of cryostorage to duckweed germplasm collections, saving labor for in vitro cultivation and maintaining characterized reference and mutant samples.
1. Introduction
The Lemnaceae (duckweeds) are the smallest and fastest growing aquatic monocotyledonous flowering plants. Due to their high adaptability to environmental conditions (temperature, pH, nutrient availability, and presence of toxic compounds), high growth rate, and capability to be dispersed by migratory waterfowl, duckweeds are widely distributed on all continents except Antarctica [1]. Due to their small size, easy maintenance, and fast vegetative multiplication, as well as the availability of published data on their physiology and biochemistry, duckweeds are considered as a model system in plant biology [2]. The potential of the practical use of duckweed is very broad; it can be used for phytoremediation of polluted waters [3], for biofuel production, for poultry and livestock feeding, for human consumption [2], and as a bioreactor for recombinant proteins [4].
The fast growth of duckweed populations is ensured by vegetative propagation, while formation of flowers and seeds occurs rarely, possibly due to the fact that some species of duckweed are interspecific hybrids [5,6]. Therefore, a range of collections of wild-type duckweed clonal isolates is maintained in different laboratories [7]. Development of genetic transformation methods [8,9,10], targeted gene editing [11], and mutant induction of duckweeds [12,13] made it necessary to preserve collections of generated transgenic and mutant duckweed lines [2].
Cryopreservation is currently considered to be the most efficient method for the long-term preservation of vegetatively propagating plant germplasm and an alternative to permanent in vitro maintenance [14]. Among cryopreservation methods, vitrification is the most used approach for plant material due to its relative simplicity and applicability to large germplasm collections, and independence from specialized controlled cooling equipment [15]. Cryopreservation by vitrification requires a preliminary increase in the cellular solute concentration often achieved by dehydration and cryoprotection of tissue to enter a glassy state during fast cooling in liquid nitrogen. During vitrification, plant tissues are dehydrated by submersing them into plant vitrification solution (PVS) to prevent the risk of ice crystal formation during cooling and warming procedures. Substances with high affinity to water molecules, such as sucrose, glycerol, and/or other polyols in high concentration, are used as major components of PVSs. These substances redistribute water by osmosis and by establishing hydrogen bounds between their hydroxyl groups and H2O molecules [16]. Dimethyl sulfoxide (DMSO) is another widely used cryoprotectant in PVS compositions. DMSO penetrates fast in to tissue [17] and increases water permeability of bio-membranes to intensify dehydration, as well as stabilizes membranes during cooling–warming and demonstrates antioxidant properties. However, DMSO may also lead to partial DNA and RNA denaturation, and to delayed cell cycle progression, enhanced Z-form of DNA, and epigenetic alterations [18]. While the composition of PVS2 and PVS1 contain DMSO in the range of 5–15% [19], PVS3 does not contain DMSO [20] and is therefore the preferred option for the subsequent study.
So far, Lemnaceae proved recalcitrant to conventional cryopreservation techniques because living in fresh water habitats makes them sensitive to desiccation stress. In addition, the duckweed fronds contain a thick layer of aerenchyma with considerable air chambers, which provide reliable buoyancy of duckweed. The surface of the air-faced side is similar to the adaxial surface of terrestrial plant leaves with stomata, cuticle, and wax layers, while the water-faced side additionally is covered with lignin/suberin layers [21], which also may impede the penetration of PVS into the cells and affect regrowth after cryopreservation.
The first successful application of cryopreservation by vitrification to duckweed accessions of Landoltia and Lemna genera was reported by Sauter (1993) [22], applying glycerol solutions of 30–60% as a cryoprotectant. Regrowth after cryopreservation was shown for 16 out of 40 tested duckweed accessions. For Spirodela, Wolffia, and Wolffiella genera, no reproducible regrowth after cooling in liquid nitrogen was achieved. The US patent WO2011005502A3 [23] uncovers a method of cryopreservation for duckweeds, which requires 1–5 weeks of pre-cultivation in a nutrition medium containing a combination of different sugars with gradually decreasing temperature and illumination and using DMSO-containing PVS for cryoprotection. Another successful cryopreservation approach for duckweed accessions has been reported recently [24]. It is based on a droplet-vitrification method using PVS2 and includes the exposure of droplets of pre-treated explants and PVS2 carried on a so-called cryo-plate to liquid nitrogen before transferring to the cryo-vials. Direct contact between the plant material and liquid nitrogen could lead to contamination [25], thus giving rise to a potential risk for successful cryopreservation of germplasm collections.
The aim of this study was to develop a simple cryopreservation method for a wide range of Lemnaceae accessions. In addition, the effects of cryopreservation on the photosynthetic performance of duckweed fronds was examined. Moreover, to elucidate the importance of the integrative regulatory hub in plant responses to low temperature, the C-repeat binding factors/dehydration-responsive element binding protein (CBF/DREB1) transcription factors was investigated. CBF/DREB1s were described as the main link regulating cold- and drought-responsive genes [26,27,28]. Therefore, CBFs play a key role in mediating the tolerance to freezing [29,30]. Plants overexpressing CBFs enhance freezing tolerance [31,32], while knockout plants become more susceptible [33,34]. Therefore, we hypothesized that a high level of CBF expression might be predictive for cell viability after cryopreservation and searched for a potential correlation between CBF gene expression during cryopreservation and the subsequent ability of duckweeds to regrow.
2. Results and Discussion
2.1. Developing the Method
Excised shoot apical meristems are the most frequently used explants for cryopreservation by vitrification. During duckweed frond development, no cytohistological zonation and/or delimitation of a tunica and corpus occur at the frond tip, indicating that no conventional shoot apical meristem is formed [35]. Potentially, axillary buds can be used as explants for duckweed cryopreservation, but excision of the explants has been published as inducing an oxidative burst, resulting in reduced viability of the plant material after cryopreservation [36]. Therefore, whole duckweed fronds were used for cryopreservation, as already proposed [22]. Portions of fronds of Le. gibba 7742 were blotted on filter paper and exposed in 1 mL of PVS3. To ensure complete and reproducible penetration of PVS3 [37] and to avoid floating of the fronds on the surface of the PVS3, we designed a special single-use insert made of aluminum foil, which allows the fronds to be kept always submersed in PVS3 (Figures S1 and S2). Incubation was for 2 h at room temperature, as employed successfully for potato, garlic, mint, and shallot [38]. To improve the penetration of PVS3 into duckweed fronds, the first 15 min of PVS3 treatment was applied under vacuum [25,39], followed by an additional 1 h 45 min at normal pressure (after vacuum release).
After the first cryopreservation experiments following the protocol given in Appendix A, the green color of the rewarmed fronds bleached and gradually turned into white when exposed to light. Even without cooling, 15 min of incubation of duckweed fronds in PVS3 at room temperature led to bleaching of the fronds after washing and subsequent illumination (12 h, 50–60 μmol·m−2·s−1) in a phytochamber for 1 day. We hypothesized that rewarmed duckweed fronds may develop daughter fronds when bleaching is delayed. Therefore, early efforts were focused on finding conditions that delay bleaching.
2.1.1. Attenuation of Toxicity of PVS3: Impact of Temperature and Illumination
A high concentration of glycerol in PVS3 (50% w/v) is reported to be toxic for many plant species [40]. We assumed that one of the key factors negatively affecting duckweed regrowth after cryopreservation is the combination of a high concentration of glycerol with light. Thus, we incubated duckweed fronds in PVS3 on ice (0 °C, general recommendation for reducing the toxic effect of a high cryoprotectant concentration) for 2 h in darkness. Unloading from PSV3 was also carried out on ice in a pre-cooled washing solution. All stages were performed in darkness. This allowed for a delay of bleaching by 5–6 days to be achieved, but without regrowth. Applying a specially designed attenuated illumination regime for regrowth (Figure S3) and gradually increasing the photosynthetic photon flux density (PPFD) and light period duration caused a delay of bleaching by up to two weeks after rewarming. However, still no regrowth was observed.
2.1.2. Addition of Antioxidants and Other Compounds That Can Support Viability
ATP [41], casein hydrolysate, amino acids, polyvinylpyrrolidone, sodium thiosulfate, silver thiosulfate, dithiothreitol, glutathione, and different combinations of growth regulators (Table S1) were tested to search for support of viability and improvement in regrowth, however, without success. Thus, the addition of antioxidants and other compounds may improve already working protocols for cryopreservation [36] but is not a key in developing a protocol for duckweeds.
2.1.3. Breakthrough Regrowth via Elevated Temperature
Cultivation under elevated temperatures (28–34 °C) is often used to rescue interspecific hybrid embryos [42]. Therefore, we checked this approach for rescuing rewarmed fronds of Le. gibba 7742 by incubation at 29 ± 1 °C during 3 weeks in a phytochamber under an attenuated light regime. Under these conditions, we observed the first regrown daughter frond derived from completely white mother fronds (Figure S4). The new fronds formed directly, without a callus stage. Overall, one regrowth event in one or two of three technical replicates of each biological replication was found and led to the use of 29 ± 1 °C in further experiments.
2.1.4. Effect of Adaptation to Cold and High Osmotic Condition at Pre-Culture Stage on Regrowth
The most common approaches for pre-culture in plant cryopreservation are cold acclimation and sucrose pretreatment, the latter considered to be more effective for plant recovery, especially for frost-sensitive species [40,43]. We tested different conditions for pre-culture of Le. gibba 7742: (1) standard (12/12 h light/dark cycle at 25 °C/22 °C); (2) cold acclimation (4 °C, darkness), and (3) imitation of night frost (16/8 h light/dark cycle at 25 °C/−1 °C, respectively), with or without increased osmotic pressure (0.015 to 0.4 M sucrose) during 6 days. Overall, pre-cultivation in a medium with 0.4 M sucrose under standard growth conditions provided the best regrowth (Table S2) compared to other conditions and, thus, was used for further experiments.
2.1.5. Illumination Regime Optimization for Early Regrowth
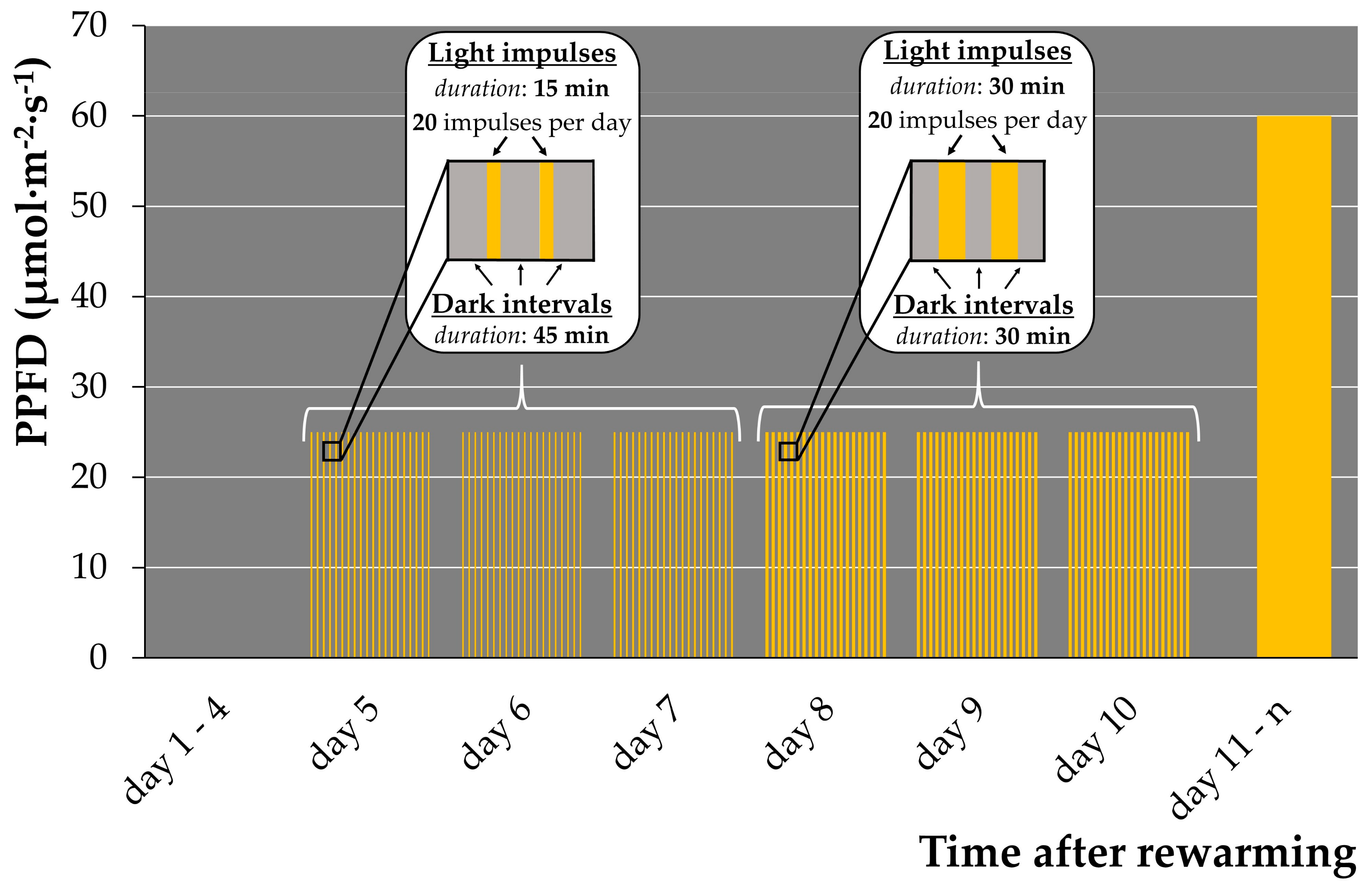
A light-sensitive Arabidopsis mutant was reported to tolerate a short-day regime with high light intensity better then a long day regime with low light intensity [44]. We previously observed that (1) rewarmed duckweeds incubated in darkness after rewarming stayed green for weeks without regrowth; (2) rewarmed duckweed exposed to attenuated light after 4 days of darkness displayed delayed bleaching but insufficient regrowth; and (3) rewarmed duckweed illuminated with a light regime of the standard growth condition bleached fast with no regrowth. Based on these observations, we hypothesized that substitution of attenuated illumination (Figure S3) by pulsed illumination of higher intensity increases regrowth. Therefore, a set of pulsed illumination regimes for the early regrowth stage with different durations of light impulses and dark intervals was applied to Le. gibba 7742 (Table 1, Figure S5).

Table 1.
Parameters of the pulsed illumination regimes for early regrowth and its impact on regrowth.
The “60/60” regime (Figure S5C) did not show differences in regrowth in comparison to the control attenuated illumination regime (Figure S3). Overall, only regimes “15/45” (Figure S5A) and “30/30” (Figure S5B) showed substantial increase in regrowth. Further, we observed two waves of appearance of new fronds: the first between days 6 and 7 after rewarming, and the second between days 9 and 11. Although more daughter fronds were generated under the “15/45” illumination regime, the duckweeds grew slower and did not proliferate “granddaughter” fronds compared with the “30/30” illumination. In addition, regrown fronds from the second wave for both illumination regimes did not form new fronds.
To ensure complete development and further multiplication of new generated fronds, we applied a combined pulsed illumination regime: four days of darkness, followed by three days of the pulsed “15/45” regime (best regrowth regime), and subsequently, three days under the “30/30” regime and transfer of rewarmed plants to standard cultivation condition (12/12 light/dark, 60 μmol·m−2·s−1) on day 11 (Figure 1). This pulsed illumination regime increased the regrowth of Le. gibba 7742 from 2–4 daughter fronds up to 9–34 per cryo-tube and ensured robust normal growth and multiplication of new fronds.
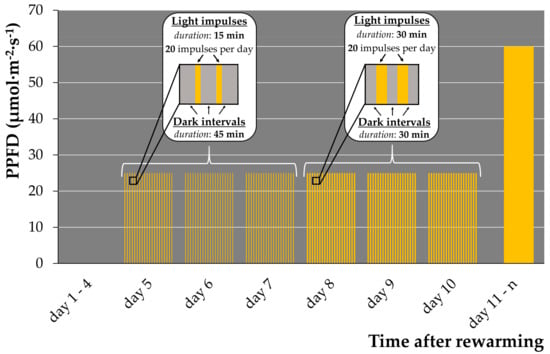
Figure 1.
Combined pulsed illumination regime used for duckweed regrowth after cryopreservation. PPFD—photosynthetic photon flux density.
The main source of damage to dehydrated green tissues under illumination is uncoupling of carbon fixation and photosynthetic electron transport chain triggering production of reactive oxygen species and, hence, oxidative stress [45,46]. At the same time, light is an important signal for plant morphogenesis and chloroplast development [47]. Therefore, the designed pulsed illumination regime provided essential light signals for induction of duckweed frond development and, consequently, regrowth. Simultaneously, the dark intervals between light impulses were sufficient for recovery of the photosynthetic apparatus and may have suppressed a burst or enabled the scavenging of reactive oxygen species.
2.1.6. Vacuum Infiltration Turned Out to Be Dispensable
Vacuum infiltration of PVS3 was used to ensure homogenous penetration of PVS3 before cooling. However, as this operation is laborious, we tested various pretreatment times with and without vacuum infiltration (Table 2) and found that the 4 h pretreatment without vacuum application produced similar results to the 4 h pre-treatment using a vacuum application. Thus, the 4 h pre-treatment in PVS3 without vacuum infiltration was used further on.

Table 2.
Duration of pre-treatment of duckweed fronds in PVS3 with and without vacuum infiltration and its impact on regrowth.
Incubation of explants in PVS on ice are recommended for many plant species [40]. The diffusion of solutions at nearly 0 °C is lower than that at room temperature; therefore, we had to increase the cryoprotection of the fronds by PVS3 up to 4 h. In addition, the saturation by cryopreservation agents of the plant tissues takes time, especially for sucrose [48]. By contrast, the cryoprotecting agent DMSO may fully penetrate leaf primordia and meristem regions within 10 min of exposure at room temperature as shown by using coherent Raman microscopy [17]. It should be noted that PVS components must not only cross the plasmalemma but also distribute throughout the whole cytoplasm volume and to desiccate cells through the redistribution of water molecules by osmosis and by establishing hydrogen bounds between their hydroxyl groups and water molecules [16]. Whatever the appropriate duration of PVS application at the cryoprotection stage is, a compromise between harmful effects of PVS and optimal desiccation of plant tissue, which is essential for successful cryopreservation, must always be found.
2.1.7. Substitution of Blotting by Spin-Drying of Duckweed Fronds before Submersing in PVS3
The considerable variation in regrowth per cryo-tube under identical conditions (Table 2) indicated that one or more factors of the cryopreservation process were not reproducible. During rewarming, we frequently noticed nontransparent white layers above frozen transparent PVS3, termed “ice caps” of varying size. These ice caps are possibly the rest of the liquid nutrition medium on the surface of duckweed fronds.
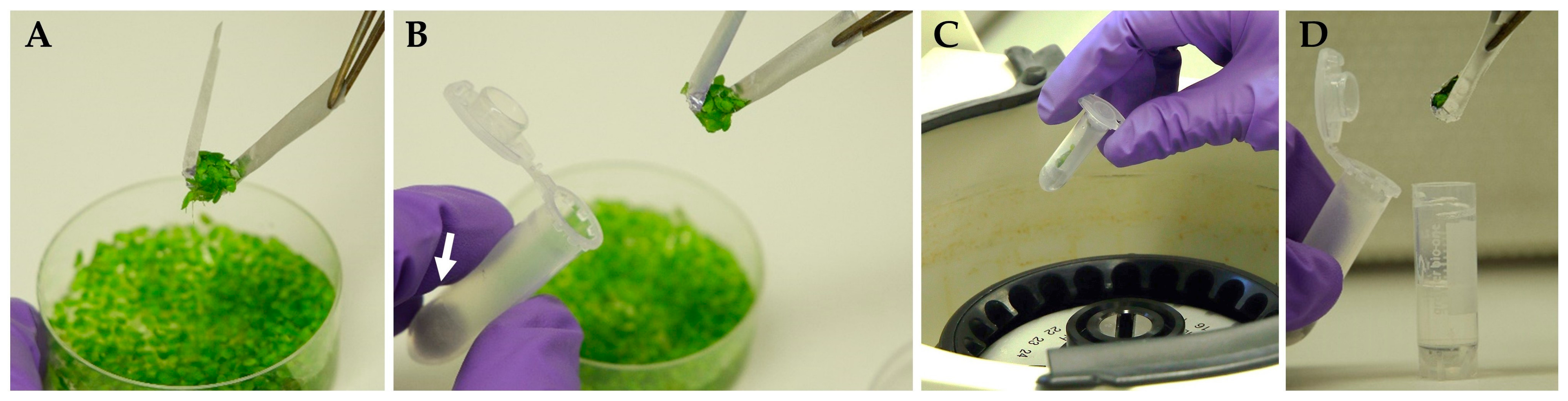
To remove the surface liquid from the aquatic plant materials, spin-drying was recommended [49]. Therefore, we modified the aluminum foil insert by making a number of tiny perforations in the middle (Figure S6) and used it to collect and to transfer duckweed fronds into 2 mL U-shaped plastic centrifuge tubes (Figure 2) with a cotton wool placed on the bottom. Centrifugation at 800× g for 3 min allowed the liquid to drain away from fronds’ surface through the perforations onto the cotton wool.

Figure 2.
Spin-drying of duckweed fronds using a perforated aluminum insert: (A) collecting duckweed fronds from culture using perforated foil insert; (B) transferring the insert with the duckweeds into the test tube; (C) spin-drying by centrifugation; (D) transferring the insert with spin-dried duckweed from centrifugal tube into a cryo-tube filled with 1 mL of PVS3. White arrow indicates a cotton wool at the bottom of the centrifugal test tube.
The perforated aluminum foil was helpful for (1) duckweed sampling and transfer of the fronds to the tube, (2) spin-drying of plant material, and (3) maintaining the duckweed fronds under the PVS3 surface in cryo-tubes during cryoprotection and cryo-cooling stages. After PVS3 vitrification using a perforated aluminum foil, the regrowth results for Le. gibba 7742 changed from 8–31 to 16–27 per cryo-tube, i.e., were less variable, and the method was faster and less laborious. Overall, the spin-drying decreased the amount of liquid remains on the duckweed surface transferred to the cryo-tubes and eliminated ice caps after rewarming.
2.1.8. Optimization of Solution Composition for Pre-Cultivation
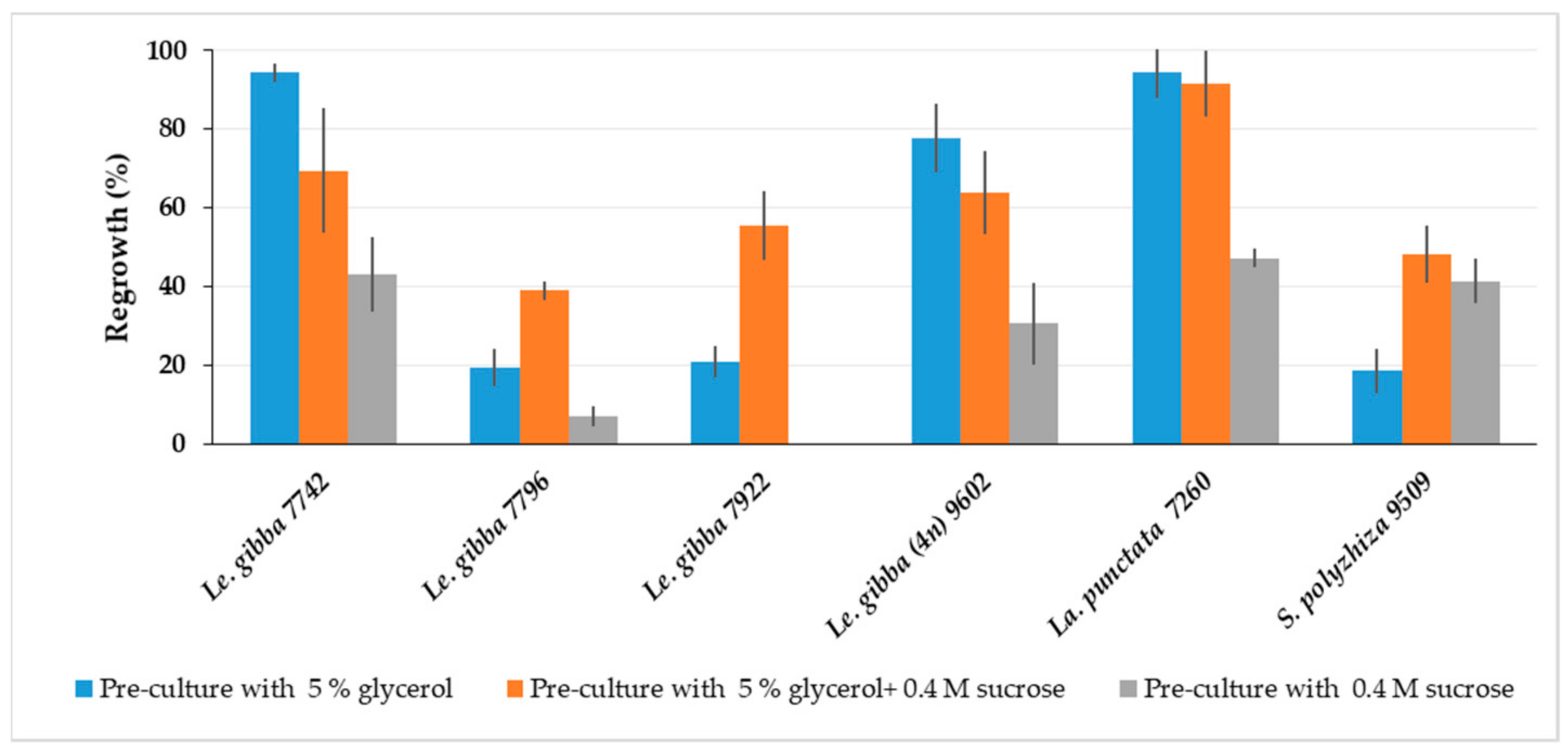
The cryopreservation protocol developed for Le. gibba 7742 using 0.4 M sucrose for pre-culture was less optimal for other duckweed accessions, presumably due to a lack of adaptation to all components of PVS3. Therefore, the pre-culture medium was supplemented with either glycerol or sucrose or both for comparison and tested for Le. gibba (7742, 7796, 7922 and 9206), La. punctata 7260, and S. polyrhiza 9509 (Figure 3). The average regrowth of duckweed cultivated in a 24-well plate was calculated as described in Section 3.7.2. Le. gibba 7742 and 9206, and La. punctata 7260 showed the best regrowth, when only 5% glycerol was applied in the pre-culture medium. For all tested duckweed accessions, except S. polyrhiza 9509, regrowth after pre-culture with 0.4 M sucrose was reduced compared to that with glycerol-containing media. Combined in a pre-culture medium, 5% glycerol and 0.4 M sucrose had a positive effect on regrowth of Le. gibba 7796 and 7922, and S. polyrhiza 9509, and other tested accessions also showed quite satisfactory regrowth; thus, this variant was chosen in further experiments.
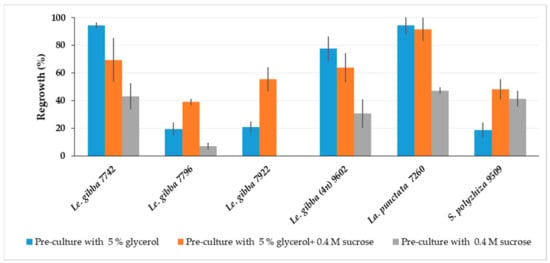
Figure 3.
Effect of pre-culture medium on regrowth (calculated according Section 3.7.2) of different duckweed accessions at day 21 after rewarming. Error bars indicate standard deviations (n = 3).
Although, the pre-culture on the medium containing both glycerol and sucrose was vital for regrowth of most tested duckweed accessions, for some of them, the pre-culture with only glycerol was preferable, probably because glycerol has a stereochemical arrangement (all the OH groups are stereochemically orientated along one side of the molecule) and small molecule size. Therefore, glycerol can interact more efficiently with the lipid bilayers and membrane proteins and can stabilize them during cryopreservation [50].
2.2. Regrowth of Spirodela, Landoltia, Lemna, and Wolffia Accessions after Cryopreservation
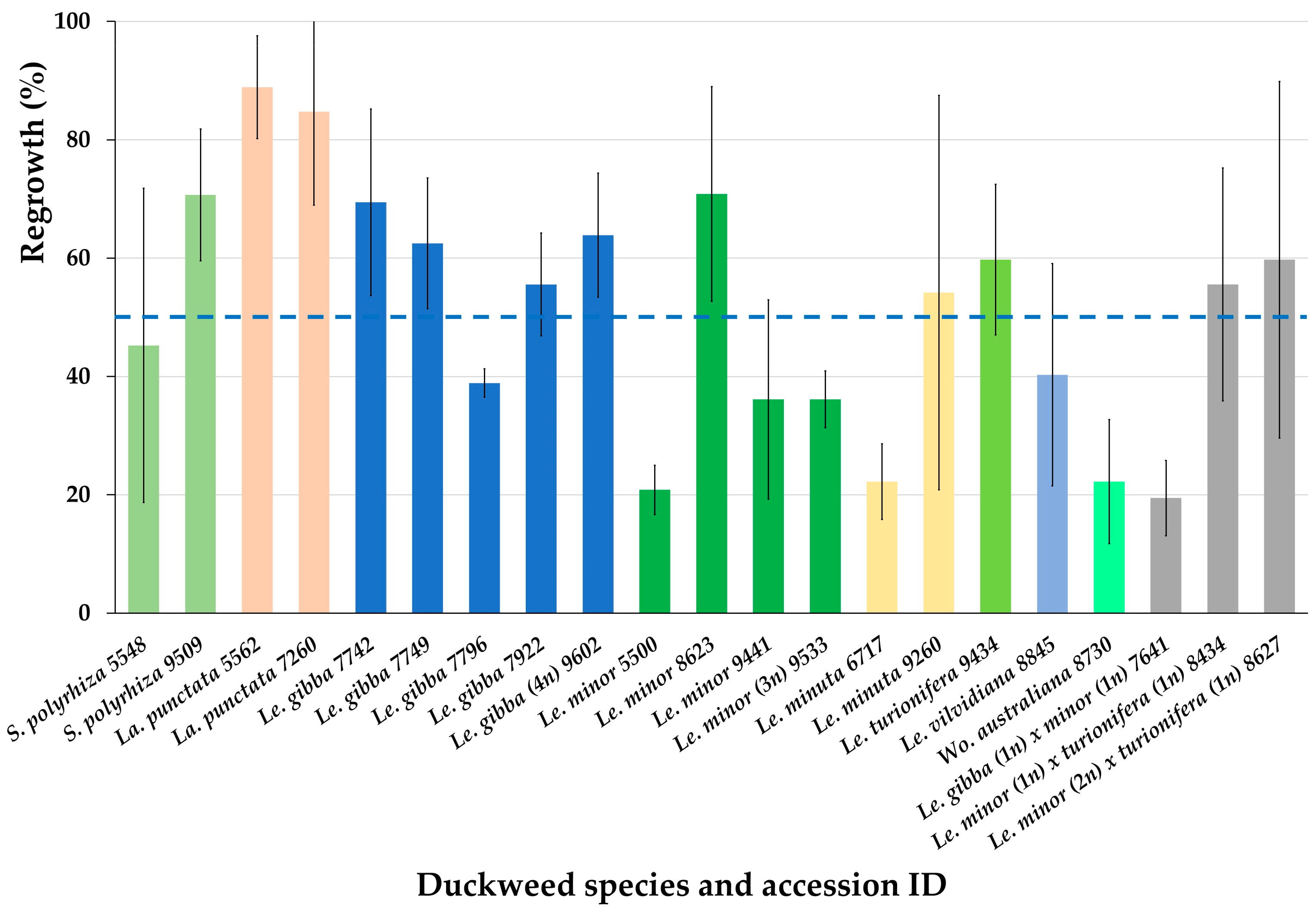
After obtaining promising regrowth results for six accession from three Lemnaceae genera (Figure 3), we cryopreserved additional 15 accessions from 4 Lemnaceae genera using the optimized protocol (Appendix B). On the average, the regrowth of all 21 cryopreserved accessions achieved 51.3 ± 20.6% (Figure 4).
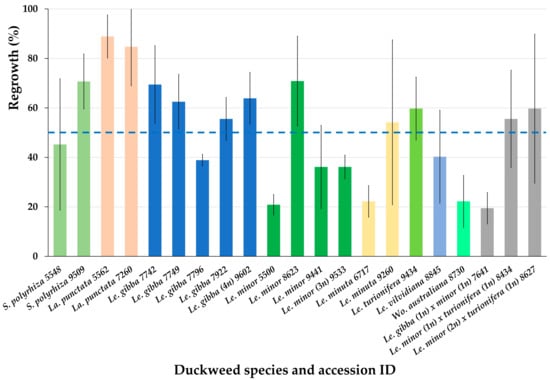
Figure 4.
Regrowth of 21 accessions from 4 Lemnaceae genera (21 days after rewarming) obtained after applying the newly developed protocol for duckweed cryopreservation. Blue line indicates average regrowth of all tested duckweed accessions. Error bars indicate standard deviations (n = 3).
Almost all tested accessions demonstrated regrowth substantially higher than 15%, the minimum viability value, established in our laboratory, and only for two of them, the regrowth was close to this cut-off value.
2.3. Flow Cytometry Demonstrates the Genome Size Stability after Cryopreservation
To estimate if the cryopreservation procedure has an impact on the genome size, measurements were carried out for donor cultures of autopolyploids Le. gibba 9206 and Le. minor 9533, as well as of interspecific hybrids Le. gibba × minor 7641, Le. japonica (minor × turionifera) 8434 and allopolyploid interspecific hybrid Le. japonica (minor × turionifera) 8627, (Table 3).

Table 3.
Genome size of polyploid and hybrid duckweed accessions before cryopreservation (donor culture) and genome size of newly developed fronds of the same accessions after cryopreservation.
Overall, no quantitative genomic changes were found between the duckweed fronds prior to cryopreservation and the newly developed fronds after cryopreservation. Regrown duckweeds were true to type according to the data from flow cytometry; therefore, the developed method for duckweed cryopreservation could be used for long-term conservation without concern for alteration in the genome size during regrowth.
2.4. Monitoring the Photosynthetic Activity of Rewarmed Fronds
The observed bleaching of duckweed fronds under illumination after rewarming raised the question about a possible correlation between functional activity of the photosynthetic apparatus in chloroplasts and the regrowth of daughter fronds. To address this question, we monitored the functional activity of the photosynthetic apparatus of rewarmed fronds during two weeks after rewarming by measuring a set of chlorophyll fluorescence parameters and subsequent calculation of photosynthetic performance in samples differing in regrowth due to different pre-cultures (see Figure 3).
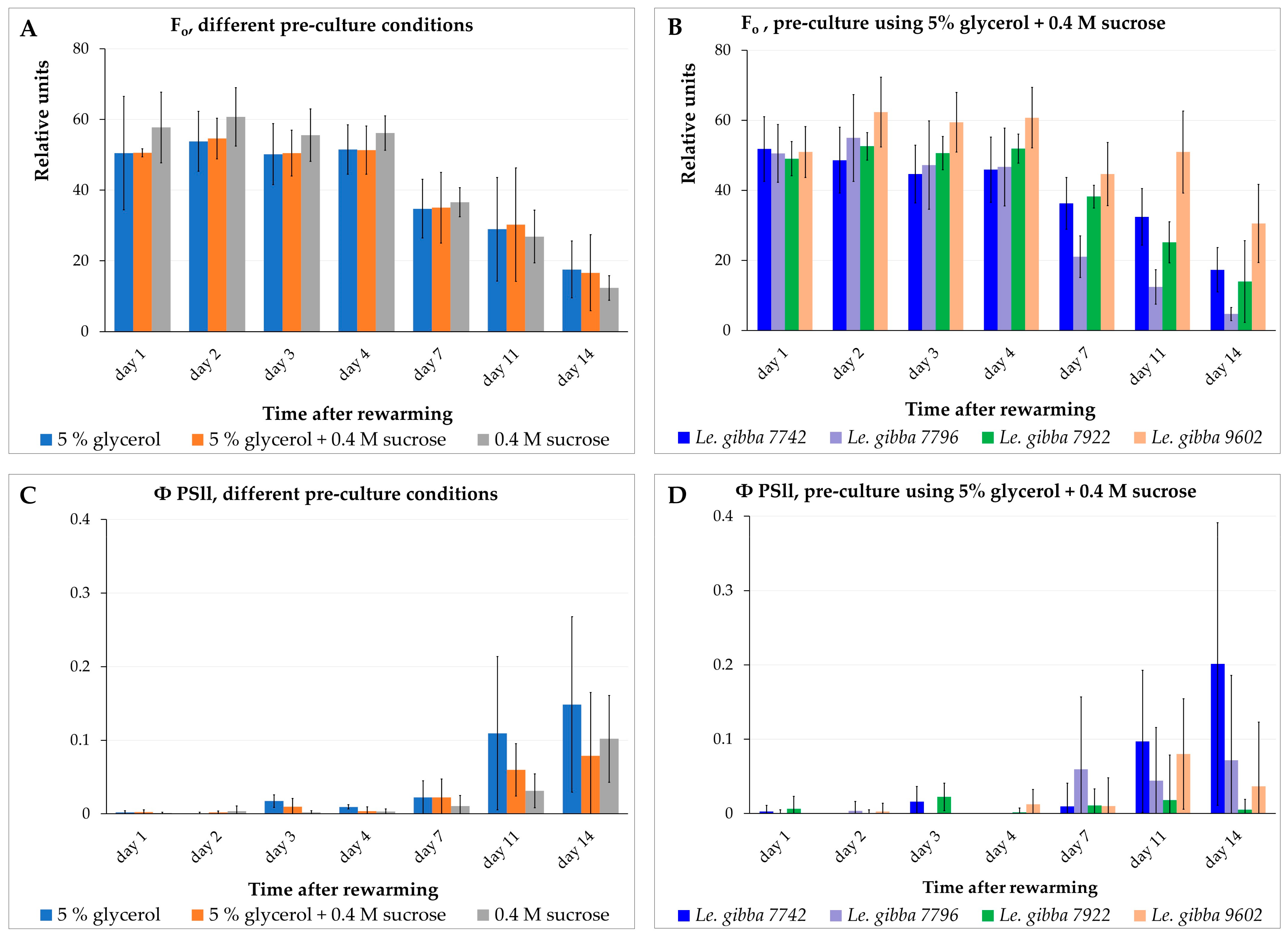
We measured the minimal fluorescence level (F0) of the dark-adapted plants, the maximal fluorescence level (Fm) transiently induced by a saturating light pulse, the maximum fluorescence yield in the light-adapted state (Fm′), and the fluorescence under actinic illumination (Fs) for the Le. gibba 7742, Le. gibba 7796, Le. gibba 7922, and Le. gibba 9602 rewarmed fronds that were pre-cultured in three different solutions before cryopreservation. The photosynthetic performance was estimated by calculating the variable fluorescence (Fv), maximum quantum yield of photosystem II (Fv/Fm), and operating efficiency of photosystem II (ΦPSII) (see Section 3.3). F0 derives directly from the pigment bed, primarily from the light-harvesting pigment antenna of PSII. In contrast, variable fluorescence Fv (difference between F0 and Fm) is closely connected to the photochemical reactions. Changes in Fv reflect mainly the redox state of the first stable electron acceptor in PSII plastoquinone A.
At the first 4 days after rewarming and cultivation in the dark, fronds displayed almost no differences in the values of F0 and Fm as well as between accessions and pre-culture conditions (Figure 5A,B and Figure S7A–C). On average, the F0 values for rewarmed materials were about half of that for the intact control material: 50.0 ± 10.1 (Figure 5A) vs. 90.0 ± 15.4 (Table S3). However, the Fm and photosynthetic performance parameters of rewarmed fronds (Fv/Fm and ΦPSII values) in all cases demonstrated values on average not more than 10% of values for intact control plants (Figure 5C,D and Figure S7G–L,N–Q, Table S3). These two observations suggest that, despite the substantial amount of chlorophyll in the rewarmed fronds, the photosynthetic machinery in the chloroplasts was almost nonfunctional and did not recover within two weeks after rewarming.
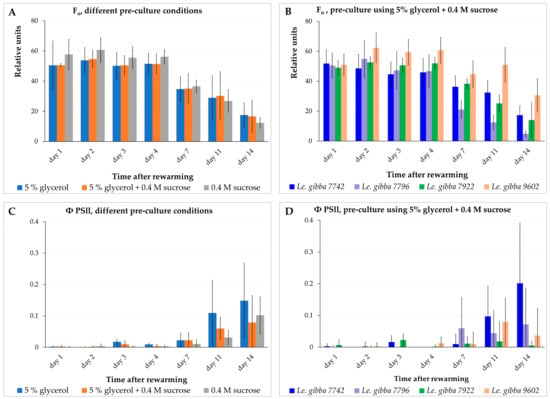
Figure 5.
Dynamics of minimal chlorophyll fluorescence F0 (A,B) and operating efficiency of photosystem II ΦPSII (C,D) of the rewarmed duckweed fronds depending on pre-culture (A,C) and duckweed accessions (B,D). For (A,C), data presented as average values for four accessions of Le. gibba (7742, 7796, 7922, 9602). Error bars indicate standard deviations (n = 3).
Between day 4 and 7 after rewarming, the F0 (Figure 5A,B) and Fm values decreased in range of 20–30% depending on accession and pre-culture solution. The observed photochemical degradation of chlorophyll was apparently the result of switching from cultivation in darkness to pulsed illumination and an elevated cultivation temperature of 29 ± 1 °C at day 5. The photosynthetic machinery did not demonstrate any signs of recovering at this period, maintaining the values of Fv/Fm (Figure S5G–I,P) and ΦPSII (Figure 5C,D and Figure S7J–L) at the same low level. From day 7 to 11, the F0 (Figure 5A,B and Figure S7A–C) and Fm (Figure S7N) values decreased less remarkably and even increased slightly for certain accessions and pre-culture conditions.
From day 5 (after switching from darkness to pulsed illumination and elevated temperature), the new daughter fronds started to grow. At this stage, the surfaces of these new tiny fronds started to overlap with the surfaces of rewarmed mother fronds; thus, a precise separation between the areas of the rewarmed fronds from areas of the new fronds was impossible during fluorescence data processing. Up to day 7, the new fronds reached larger dimensions and developed their photosynthetic apparatus. Thus, subsequent decreasing of the F0 (Figure 5A,B and Figure S7A–C) and Fm (Figure S7N) values of the rewarmed fronds was partially or completely compensated by increasing of chlorophyll fluorescence of the new daughter fronds. The increasing Fv, Fv/Fm, and ΦPSII values at this stage for most accessions (Figure S7D–L) and pre-culture conditions (Figure S7O–Q) also point to a possible impact of newly developed fronds on the results of fluorescence measurements.
From day 11 to 14, the F0 and Fm values (Figure 5A,B and Figure S7A–C,N) decreased by half, apparently due to switching from cultivation under pulsed illumination and elevated temperature (29 ± 1 °C) to the standard condition (usual diurnal illumination and temperature 26/25 °C, 12 h/12 h light/dark period) on day 11 and further degradation of chlorophyll in rewarmed fronds and/or because a part of the tiny daughter fronds might have died or their development was interrupted. This assumption is supported by the observation that even the Fv values calculated for fronds that did not generate visible daughter fronds increased slightly from day 7 to 11 and decreased again after the 11th day for most of the accessions and pre-culture conditions. At this stage, differences regarding the F0 and Fm values between individual accessions appeared, suggesting that further individual optimization of switching between illumination regimes might be possible for some accessions.
The photosynthetic performance of the rewarmed fronds remained—with minor variations—at a low level during the first week of the observation (Figure 5C,D and Figure S7D–L,O–Q). This indicated that the photosynthetic apparatus of the fronds was irreversibly damaged during cryopreservation.
The Le. gibba accessions displayed in this experiment substantially different regrowth after cryopreservation (Figure S7A–L) depending on the composition of the pre-culture solution (Figure 3). However, the values of the chlorophyll fluorescence and photosynthetic performance of the rewarmed fronds, averaged by values for individual accessions and grouped by the type of pre-culture solution, demonstrated no substantial differences, neither in the level nor in their dynamics during the first week after rewarming. This indicates an absence of a correlation between the photosynthetic performance of the rewarmed fronds and regrowth after cryopreservation.
In numerous studies [53,54,55,56], dismantling thylakoid membranes and plastoglobuli formation is demonstrated for plant tissue pretreated with PVS. Chlorophyll is also degraded upon dehydration stress [45,57]. Despite the apparent presence of chlorophyll in fronds right after rewarming in darkness, the photosynthetic apparatus lost its function, as we showed by monitoring the efficiency of photosystem II. Non-functioning duckweed plastids are probably not able to recover their structure and function, and the chloroplasts of regrown plants evolved from proplastids of meristem cells [58]. Duckweed regrowth occurred only via de novo development of daughter fronds from meristems located in reproductive pockets, whereas green mother fronds were not able to survive during cryopreservation procedure. Our data confirm the statement that only meristematic cells from apical dome and youngest leaf primordia can survive after cryopreservation [59].
2.5. Identification of Duckweed CBFs and Their Involvement in Cryopreservation
2.5.1. Identification and Characterization of CBF/DREB1 Genes in Duckweeds
DREB protein subfamily members contain a highly conserved APETALA2/ethylene-responsive element-binding factor (AP2/ERF) domain with a characteristic valine residue at position 14 that determines the binding affinity to cis-regulatory dehydration-responsive elements (DRE), distinguishing them from related ERF transcription factors [60,61]. Using the tBlastN program, we identified 21 putative genes from the genome of Le. gibba, 28 genes of La. punctata, and 20 S. polyrhiza-encoding proteins with a DREB-specific domain (Figure S8). These protein sequences, 56 DREB proteins from Arabidopsis and 50 DREB proteins from rice, were taken for phylogenetic analysis. The collected DREBs were clustered into four groups (I–IV, Figure S9). Since Group A1 (CBF/DREB1) has been described to be involved in cold and freezing tolerance, a focus was taken on the CBF/DREB1 representatives of the duckweed genomes. The phylogenetic reconstruction showed that three CBFs of Le. gibba and two each of La. punctata and S. polyrhiza clustered in Group A1 (CBF/DREB1) together with the Arabidopsis CBFs AtCBF1-4. The identified CBFs are highly similar and form an individual subtree. The identified proteins from Le. gibba, La. punctata, and S. polyrhiza, we designated as LgCBF1, LgCBF2, LgCBF3, LpCBF1, LpCBF2, SpCBF1, and SpCBF2. Besides the AP2/ERF domain, which is involved in DNA binding, the CBFs possess the LWSY-motif in the activation domain at the C-terminus and a nuclear localization signal (NLS) near the AP2/ERF domain (Figures S8 and S10). Unlike AtCBF1-3, which forms a self-regulatory cluster, duckweed CBFs are located at a large distance from each other (LgCBF2 and LgCBF3) and even on different chromosomes.
To gain insight into the transcriptional regulation of CBF/DREB1 genes, we analyzed their promoter regions for the presence of regulatory cis-elements. The survey revealed both E- and G-boxes in the duckweed CBF promoters that indicates the possibility of regulation of their expression by low temperature and light (Table S4). All promoters of duckweed CBF genes contained multiple elements responsive to water stress and dehydration (MYC) as well as to methyl jasmonate, abscisic acid (ABRE), and ethylene (ERE). Compared to Arabidopsis, the promoters of duckweed CBF genes have reduced numbers of cis-elements for the circadian rhythm of expression.
Numerous experimental data have demonstrated that the expression of CBFs is regulated by a circadian clock in Arabidopsis [62,63]. Therefore, we measured the mRNA abundance of LgCBF1-3 in Le. gibba 7742 every 4 h during a day and revealed no oscillation of mRNA abundance for LgCBF1, whereas LgCBF2 and LgCBF3 showed circadian regulation with a peak at 4:00 corresponding to the Zeitgeber unit (Figure S11). The relative mRNA abundance increased about 4- and 12-fold for LgCBF2 and LgCBF3, respectively, indicating also a dependency of LgCBF2 and LgCBF3 on the circadian clock in duckweed. However, the diurnal rhythm in duckweed was expressed weaker than was reported for Arabidopsis [62,63].
The mRNA abundance of LgCBF1-3 in response to cold and osmotic stress treatment (Figure S12), usually applied during the pre-culture step, was analyzed. Exposition of Le. gibba 7742 to +4 °C for 3 days resulted in an increase in LgCBF1-3 mRNA abundance: the detected relative increase was about 28-, 103- and 131-fold, respectively, compared to the control condition (+25 °C). After the addition of 0.4 M sucrose, all tested genes were induced and showed elevated transcript levels, especially for LgCBF1, with an expression 80-fold higher than that under the control condition (SH, 4 °C). The highest mRNA abundance was recorded for LgCBF1 after treatment with 0.4 M sucrose at 4 °C.
2.5.2. Dynamics of CBF mRNA Abundance during Cryopreservation
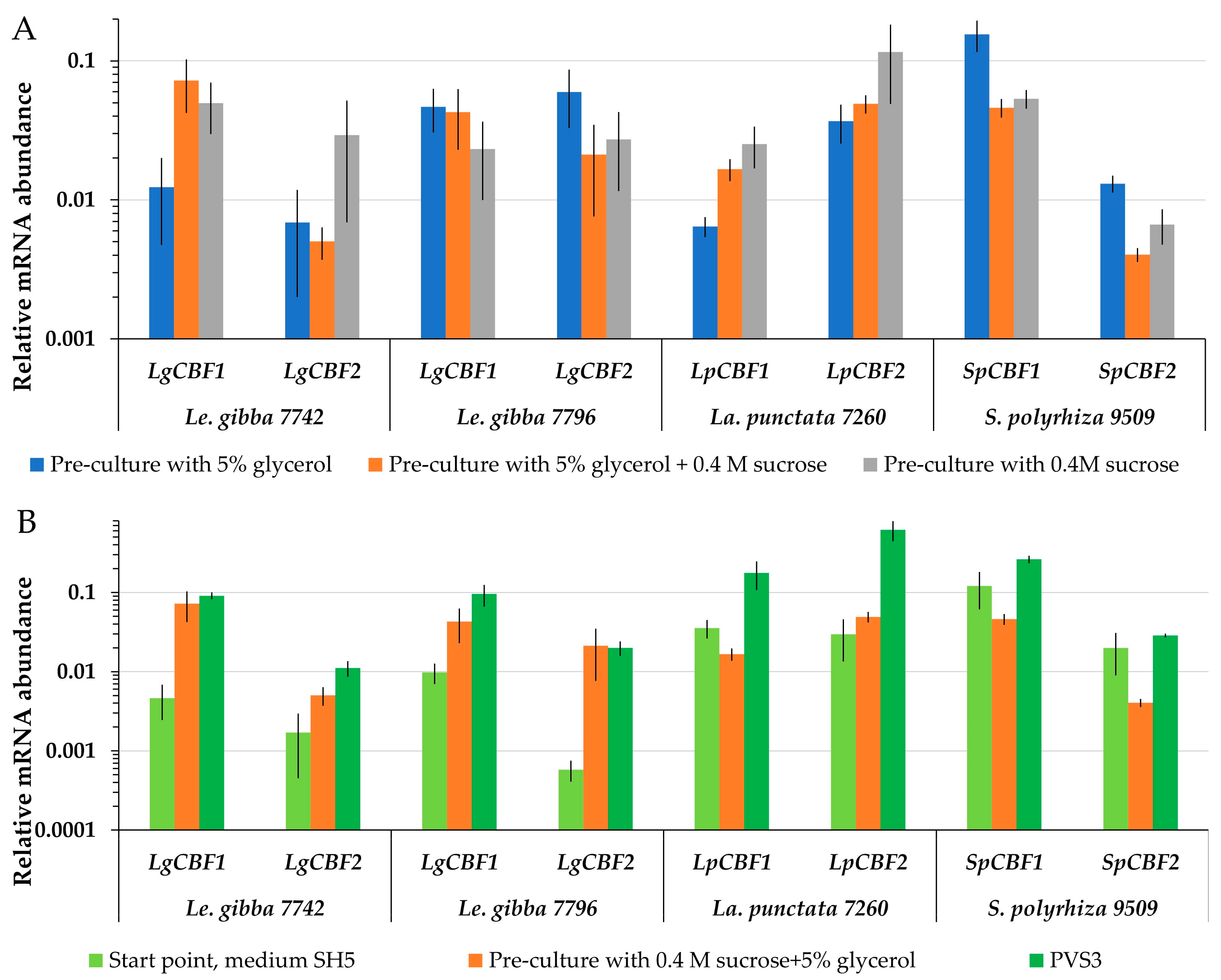
Searching for a potential link between the expression of CBF genes during cryopreservation and the ability to re-grow, the mRNA abundance of CBF1 and CBF2 turned out to be highly variable. The level of LgCBF1 mRNA abundance of Le. gibba 7742 was the highest after pre-culture with 5% glycerol and 0.4 M sucrose (Figure 6A), and LgCBF2 was more abundant after application of 0.4 M sucrose during pre-culture (Figure 6A). However, the highest regrowth of Le. gibba 7742 was observed after pre-culture of fronds with 5% glycerol (Figure 3). For Le. gibba 7796 and S. polyzhiza 9509, the highest mRNA abundance of CBF1 and CBF2 was recorded after pre-culture in 5% glycerol. Unlike for Le. gibba 7796, the regrowth efficiency of S. polyzhiza 9509 was higher after pre-cultivation with 5% glycerol and 0.4 M sucrose (Figure 3). The mRNA abundance of the LpCBF1 and LpCBF2 genes of La. punctata 7260 was the highest after pre-cultivation in 0.4 M sucrose, which resulted in the lowest regrowth. Thus, the mRNA abundance of CBF in duckweed fronds did not depend on the pre-culture conditions.
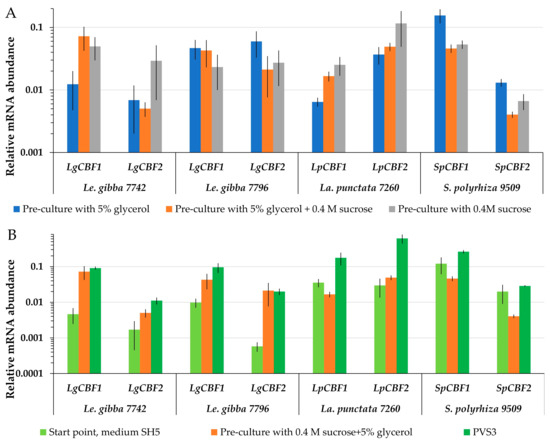
Figure 6.
Relative mRNA abundance of duckweed CBF1 and CBF2 genes (A) after different pre-culture treatment 5% glycerol and/or 0.4 M sucrose (see Figure 3 for corresponding regrowth data); (B) during preparation of duckweed to cryopreservation, including PVS3 treatment. The start points were used as controls. The relative mRNA abundance was normalized against the mRNA abundance of histone H3 (LgH3). Error bars indicate standard deviations (n = 5).
PVS3 triggered the accumulation of CBF. The final protocol of duckweed cryopreservation included the cultivation on SH medium followed by pre-culture in 5% glycerol plus 0.4 M sucrose and 4 h incubation in PVS3. During the pre-culture step, the CBF1 and CBF2 mRNA abundance was increased in Le. gibba 7742 and 7796 compared to the start point (Figure 6B). The following incubation in PVS3 yielded a further increase in LgCBF1 mRNA abundance. Transferring of La. punctata 7260 fronds to the pre-culture solution resulted in a slight decrease in LpCBF1 mRNA abundance and an increase in LpCBF2, while mRNA abundance of both genes increased significantly after incubation in PVS3. Pre-culture of S. polyzhiza 9509 with 5% glycerol and 0.4 M sucrose reduced the levels of CBF1 and CBF2, whereas the application of PVS3 increased them in relation to the start point. For all accessions, the regrowth after cooling and relative mRNA abundance of CBF genes as well as their dynamic patterns during preparation of the duckweed to cryopreservation were not correlated.
CBFs were reported in numerous publications to play a pivotal role in cold acclimation and acquisition of freezing tolerance [28,29,30,31,32,33,34]. In Arabidopsis thaliana, 12% of the cold-responsive genes are controlled by the CBF pathway, and approximately, 10% of the cold-activated genes are regulated by the HY5 pathway [64]. Thus, the CBF pathway is not the only one essential for cold response but also, in duckweed, other stress response pathways may be activated. Moreover, the expression of CBFs is highly regulated at many levels including transcription, mRNA stability, translation, post-translational modifications, and protein turnover. Recent studies [65] demonstrated that Arabidopsis CBF is activated by a redox-dependent switch. Only CBF monomers reduced by Thioredoxin h2 can induce cold-regulated gene expression, indicating the importance of the cellular redox potential in cold response. The mRNA abundance of CBF showed no correlation with the frequency of duckweed regrowth; thus, it cannot be used as a marker for further optimization of the cryopreservation procedure.
3. Materials and Methods
3.1. Plant Material and Growth Conditions
Duckweed accessions used in this study are from a stock collection at the Matthias Schleiden Institute—Plant Physiology, University of Jena, and were kindly provided by Klaus Appenroth (Table 4). Duckweed cultures were aseptically maintained on a liquid Schenk and Hildebrand (SH) nutrition medium, pH 5.8 (Duchefa S0225, Haarlem, NL, USA), supplemented with 5 g/L sucrose (medium SH5) in a phytochamber set at 12/12 h light/dark cycle, at a PPFD of approximately 60 μmol·m−2·s−1 with the spectrum shown in Figure S13, and at 26 °C/25 °C for the light/dark periods. For Wolffia australiana 8730 cultivation, a liquid SH medium, supplemented with 5 g/L sucrose, 0.5 g/L casein hydrolysate, and 0.5 g/L yeast extract (pH 5.5), was applied. Before the cryopreservation, the duration of the last subculture of donor duckweed plants was 1–2 weeks.

Table 4.
The list of duckweed species, polyploids, and hybrids used for cryopreservation.
For cold acclimation (see Section 2.1.4) of the duckweeds during the 6-day pre-culture prior to cryopreservation, we used +4 °C in a refrigerator or a phytochamber set in the 12/12 h light/dark cycle, at a PPFD of approximately 60 μmol·m−2·s−1 with the spectrum shown in Figure S13, and at 20 °C/1 °C for the light/dark periods.
For chlorophyll fluorescence measurement (see Section 2.4), duckweed fronds of different accessions Le. gibba were pre-cultured in solutions of different composition according to Section 2.1.8. For each accession and each pre-cultivation condition, 3 cryo-tubes (as 3 technical repeats) with 25–50 separated fronds or unseparated colonies in each cryo-tube were cryopreserved. Rewarmed fronds were washed in 1.2 M sucrose solution (for 1 h, on ice, in darkness) and transferred to 0.9% glucose solution in darkness at 25 °C for further washing and revitalization. The day after rewarming and washing, 8 rewarmed separated fronds or unseparated colonies from each of three cryo-tubes of the same accession and pre-cultivation condition (8 × 3 = 24 fronds or colonies) were randomly selected and transferred to plastic Petri dishes, (Ø 9 cm) on a SH solid nutrition medium, supplemented with 0.5% sucrose, 0.5% glucose, and 1% agar (pH 7.1–7.2).
For diurnal rhythm analysis of CBF gene expression (see Section 2.5.1), 21 flasks were inoculated with Le. gibba 7742 and grown in sugar-free SH medium for 4 weeks; then, the medium was replaced with a fresh one of the same composition for another 3 days. Three independent biological replicates were randomly sampled every 4 h. The first samples were collected at 8:00, when the lighting in the phytochamber started (0:00, 4:00, 8:00, 12:00, 16:00, 20:00, and 0:00 corresponding to Zeitgeber unit).
For the gene expression analysis during preparation of the duckweed for the cryopreservation procedure (see Section 2.5.2), duckweed fronds were collected (1) before immersion of the fronds to pre-culture solution (the start point), (2) at the end of pre-culture stage, and (3) at the end of exposition in PVS3 prior to cryo-cooling in liquid nitrogen.
3.2. Nuclear Genome Size Measurement
Genome size measurements were performed according to [66] using a CyFlow Space flow cytometer (Sysmex-Partec GmbH, Münster, Germany). Nuclei were isolated by chopping fronds of the duckweed clones with a sharp razor blade together with young leaf tissue of Lycopersicon esculentum Mill. convar. infiniens Lehm. var. flammatum Lehm. ‘Stupicke Rane’ (IPK genebank accession number: LYC 418, DOI: 10.25642/IPK/GBIS/53282) as an internal reference standard using the DNA staining kit ‘CyStainR PI Absolute P’ (Sysmex-Partec GmbH, Münster, Germany). Approximately 10,000 nuclei per sample were analyzed, and at least four independent measurements per clone were performed on two independent days. The absolute DNA contents (pg/2C) were calculated based on the mean values of the G1 peak and the corresponding genome sizes (Mbp/1C) according to [67].
3.3. Chlorophyll Fluorescence Measurement
Chlorophyll fluorescence parameters were measured using the pulse-amplitude-modulated (PAM) technique [68,69,70] and the FluorCam device (Photon Systems Instruments, Brno, Czech Republic) installed in an automated phenotyping platform [71]. Measurements were performed for days 1, 2, 3, 4, 7, 11, and 14 after rewarming.
In the dark-adapted state, the minimal fluorescence level (F0) was determined by applying a weak, pulsed measuring light (PAR ≤ 0.2 µmol photons m−2 s−1) that does not drive photosynthesis, and a saturating light pulse (800 msec; PAR: 3600 µmol photons m−2 s−1) was applied to induce the transiently maximal fluorescence level (Fm).
Variable fluorescence (Fv) was calculated as follows:
Fv = Fm − F0
The maximum quantum yield of PSII is given as Fv/Fm.
Subsequently, induction of chlorophyll fluorescence was followed for 3 min at a light intensity of 100 µmol photons m−2 s−1. Finally, the maximum fluorescence yield in the light-adapted state (Fm′) was measured during exposure to a saturating light flash, and the operating efficiency of PSII (ΦPSII) was determined from the steady state chlorophyll fluorescence under actinic illumination (Fs) and Fm′ according to the following equation:
ΦPSII = (Fm′ − Fs)/Fm
The fluorescence data were processed using FluorCam7 software (version 1.2.5.24, Photon Systems Instruments, Brno, Czech Republic) and subsequently were exported to MS Excel for further calculation and charts drawing. During fluorescent data processing for each separated frond or unseparated colony, the individual areas were selected manually with subsequent background exclusion using a cut-off value of 20. The areas corresponding to new daughter fronds which appeared after rewarming were manually (as precise as possible) excluded from the areas of rewarmed mother fronds.
The measured values of the fluorescence parameters of each individual frond or colony were averaged for each sample by all rewarmed 24 fronds or colonies in each Petri dish (that represented 8 fronds randomly taken at the second day after rewarming from 3 cryo-tubes).
3.4. Identification and Analysis of CBF/DREB1 Gene Family Members of Duckweeds
To access the duckweed orthologues of genes encoding transcription factors of the CBF/DREB1 family, we searched the duckweed genome sequences using as queries the reference protein sequences of AtCBF1 (At4g25490, NP_567721.1), AtCBF2 (At4gc25470, NP_567719.1), and AtCBF3 (At4g25480, NP_567720.1) from the dicotyledonous model plant Arabidopsis thaliana and from the monocotyledonous model plant Ozyza sativa OsDREB1A (Os09g35030, AAN02486.1), and OsDREB1B (Os09g35010, NP_001409784.1), OsDREB1C (Os06g03670, NP_001407946.1), downloaded from the NCBI database [72]. The tBlastN program was used to identify DREB gene subfamily members in genomes of Le. gibba 7742a (v0.5.1, id25249), La. punctata 5635 DWC138 (v2, id63586), and S. polyrhiza 9509 (voxford_v3, id51364) available on the CoGe Comparative Genome Browser [73]. To verify the search results, we analyzed all candidate sequences using InterPro [74]. The conserved motifs in CBF/DREB1 protein sequences were found using the MEME tool [75]. Multiple alignments and phylogenetic reconstructions were performed using the function “build” of ETE3 3.1.2 [76], as implemented on the GenomeNet [77]. Alignment was performed with MAFFT v6.861b with the default options [78]. The ML tree was inferred using IQ-TREE 1.5.5 ran with ModelFinder and tree reconstruction [79]. A best-fit model according to BIC was VT+F+R7. Tree branches were tested by SH-like aLRT with 1000 replicates. To address potential cis-regulatory elements in the promotor sequences of the duckweed CBFs, the sequences 2000 bp upstream from the start codon of the CBF genes were extracted from the genome database and analyzed using the PlantCARE tool [80].
3.5. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time RT–PCR
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany) from 100 μg of duckweed fronds, which were collected, thoroughly blotted, frozen in liquid nitrogen, and then ground using a RNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany). The quantity of isolated RNA was estimated using a NanoDrop One C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). In total, 500 ng of total RNA was used as a template for synthesis of the first cDNA strand, using RevertAid H Minus First Strand cDNA Synthesis Kit, primed by oligo(dT)18 (Thermo Fisher Scientific, Waltham, MA, USA), in a total volume of 20 µL and diluted to 1:5 with nuclease-free water. qRT–PCR was performed in a 384-well thermocycler QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems™, Waltham, MA, USA) using the PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Darmstadt, Germany) and gene-specific primers (listed in Table S5). Four to five identically treated biological replicates with three technical repeats were analyzed. The resulting data were analyzed using QuantStudio™ Real-Time PCR Software v1.1 (Applied Biosystems™, Waltham, MA, USA) employing corresponding histone H3 and β-actin genes as reference genes (primer sequences are given in Table S5). The level of relative expression was calculated using the 2−ΔΔCt method [81] and visualized using the Microsoft Excel 2016 program.
3.6. Measurement of Spectral Characteristic of the Light Source in the Phytochamber
The relative spectral power distribution of light from the light sources in the phytochamber, used in experiments, was measured using the HPCS300P Spectral Illuminance Sensing Module (Hangzhou Hopoo Light and Color Technology Co., Hangzhou, China). The data processing was performed using OHSP Spectral Illuminance Analyzer V 1.42 software (Hangzhou Hopoo Light and Color Technology Co., Hangzhou, China).
3.7. Statistical Analysis
All experimental conditions were reproduced independently a minimum of 3 times as biological replicates. Each biological replicate included 3 technical repeats as 3 cryo-tubes, the duckweeds of which were independently processed after rewarming. Using whole duckweed fronds submersed in PVS3 as the material for cryo-cooling and due to the relatively small dimensions of duckweed fronds, we did not calculate the exact number of fronds that we transferred to the cryo-tube.
3.7.1. Optimization Experiments
The number of Le. gibba 7742 fronds transferred to the cryo-tubes during optimization experiments (25–50 fronds per cryo-tube) was calculated by visual analysis of 40 representative photo-images of the rewarmed fronds, transferred from one cryo-tube to a glass tube with liquid nutrition medium after rewarming. To accelerate and simplify the collection and processing of the results of the optimization experiments, the regrowth events per cryo-tube, observed for all 3 biological replicates and the technical repeats for each experimental condition, were visually calculated and provided as a range from absolute minimal to maximal values.
3.7.2. Calculation of Average Regrowth
The average regrowth was calculated for the experiment aimed at optimizing the solution composition for pre-cultivation for all tested duckweed accessions cryopreserved using the optimized cryopreservation protocol and for experiments of CBF1-2 expression and chlorophyll fluorescence measurements. The regrowth was calculated for 24 separated duckweed fronds (or colonies) per experimental condition, three in each well of a 24 well plate filled with SH medium and supplemented with 0.5% sucrose and 0.5% glucose (pH 7.1–7.2). The three times 8 wells represented 3 technical repeats (corresponding to 3 cryo-tubes). Each well of the plates was photo-documented for subsequent regrowth calculation. The resulting regrowth of each well can take one of four values: 0—no regrowth event detected in the well; 1—one rewarmed mother frond or unseparated colony generated (a) new frond(s); 2—two rewarmed mother fronds or unseparated colonies generated new frond(s); and 3—three rewarmed mother fronds or unseparated colonies generated new frond(s). The values for the regrowth for each of the 8 wells with the fronds from one technical repeat were summarized (representing the number of regrowth events for a given technical repeat) and were divided by 24 (the whole number of monitored rewarmed fronds from a given cryo-tube). To obtain the average regrowth, the mean values and standard deviation were calculated for all biological replicates for each used duckweed accession and/or experimental condition. All calculations were performed using MS Excel software from Microsoft Office Standard 2019. The standard deviation values were calculated using the “STDEV.S” function.
4. Conclusions
A simple, fast, and DMSO-free protocol for cryopreservation by vitrification of duckweed was developed. The protocol was successfully tested on a wide range of Lemnaceae accessions of the genera Spirodela, Landoltia, Lemna, and Wolffia. Cryopreservation of duckweed did not cause obvious changes in the genome size of polyploids or interspecific hybrid accessions. Using whole duckweed fronds instead of excised meristems for cryopreservation saves time and labor and allows for rapidly changing conditions for optimization. To our best knowledge, the representatives of the Lemnaceae family are the first freshwater angiosperms being successfully cryopreserved and, therefore, could be considered as the basic model for the cryopreservation of vascular freshwater aquatics.
A combination of pulsed illumination with elevated cultivation temperature for the early regrowth stage yielded for most tested duckweed accessions a substantial increase in regrowth that was in all cases higher than the minimal viability criteria of 15%. Using a newly designed single-use perforated foil insert allowed for fast and less laborious sampling of duckweed fronds from the donor culture, providing increasing protection against contamination, especially when simultaneously working with a large number of accessions, and decreasing the variability in regrowth results by spin-drying of duckweed fronds from the rests of liquid nutrition medium before submersion in PVS3.
Despite a substantial amount of chlorophyll in the rewarmed fronds, their photosynthetic machinery in the chloroplasts was not functional and did not recover. A green color of the rewarmed fronds and chlorophyll fluorescence by itself cannot be considered for viability. However, monitoring of photosynthetic performance via measurement of the chlorophyll fluorescence parameters of rewarmed fronds can serve as sensitive and non-destructive real-time tool for the detection and estimation of early regrowth of new fronds emerging from rewarmed fronds after cryopreservation. Moreover, this method can point out potentially non-optimized elements in the protocol for early regrowth.
The mRNA abundance of CBF during the preparation of duckweed for cryopreservation cannot be used as a genetic marker for viability and regrowth competence after cryopreservation. Probably, the CBFs are necessary for adaptation to various abiotic stress appearing during cryopreservation but are insufficient to predict successful regrowth of duckweed after cryopreservation.
Overall, our novel cryopreservation protocol (Appendix B) provides the basis for future routine cryopreservation of duckweed germplasm collections. The outcome of our optimization experiments could be useful in developing cryopreservation for other, in particular aquatic, plant species. Further testing is necessary to expand the protocol for cryopreservation to all species of the Lemnaceae family.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants12183302/s1, Figure S1: Preparation of the “U-shaped” aluminum foil insert; Figure S2: Preparation of duckweed frond portions for cryopreservation using the “U-shaped” aluminum foil insert; Figure S3: The scheme of the attenuated illumination regime; Figure S4: Representative image of the regrowth of a new (daughter) fronds from rewarmed (mother) fronds of L. gibba 7742; Figure S5: Graphical scheme of the pulsed illumination regimes; Figure S6: Preparation of perforated aluminum foil inserts; Figure S7: Dynamics of chlorophyll fluorescence parameters and photosynthetic performance of rewarmed fronds at early regrowth stage; Figure S8: Structure of identified CBF/DREB1 proteins in Le. gibba, La. punctata, and S. polyrhiza compared with homologues from Arabidopsis and rice; Figure S9: Phylogenetic tree of DREB protein subfamily members from Arabidopsis, rice, Le. gibba, La. punctata, and S. polyrhiza; Figure S10: Sequence alignment of CBF/DREB1 proteins from Arabidopsis, rice, Le. gibba, La. punctata, and S. polyrhiza; Figure S11. Relative mRNA abundance of Le. gibba 7742 LgCBF1, LgCBF2, and LgCBF3 during daytime, relative to histone H3 (LgH3); Figure S12. Relative mRNA abundance of Le. gibba 7742 LgCBF1, LgCBF2, and LgCBF3 genes dependent on cold and osmotic treatment, normalized against mRNA abundance of histone H3 (LgH3); Figure S13: Relative spectral power distribution of light from light sources in climate chamber, used in experiments; Table S1: Effect of supplemented compounds used in different solutions on cryopreservation results of Le. gibba 7742 after rewarming; Table S2: Regrowth of Le. gibba 7742 depends on pre-culture condition; Table S3: Parameters of chlorophyll fluorescence and photosynthesis performance for intact Le. gibbba 7742 culture; Table S4. Distribution of potential regulatory DNA cis-elements in promoter region of CBF genes in A. thaliana and Le. gibba 7742; Table S5: List of RT-qPCR primers used for evaluating expression of the CBF genes in Le. gibba, La. punctata, and S. polyrhiza.
Author Contributions
Conceptualization, A.P., O.K., M.K., I.S. and M.N.; methodology, A.P., O.K., N.T., H.T., J.F. and M.N.; validation, I.S. and M.N.; formal analysis, I.S. and M.N.; investigation, A.P., O.K., M.K., J.F. and H.T.; resources, M.N. and M.K.; data curation, M.K., H.T., J.F. and M.N.; writing—original draft preparation, A.P., O.K., H.T. and J.F.; writing—review and editing, M.K., H.T., J.F., N.T., I.S. and M.N.; supervision, M.N. and I.S.; project administration, M.N.; funding acquisition, A.P., M.N. and I.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Philip Schwartz Initiative of the Alexander von Humboldt Foundation, grant number 0901324; also partially by VolkswagenStiftung, grant number Az.: 9C069; and partially by the concept phase of EURODUCKWEED, BMFB grant# 100632287. Costs for open access publishing were partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant 491250510).
Data Availability Statement
All the data generated in this work are accessible in the main text of the article and its Supplementary tables, figures, and Appendix A and Appendix B.
Acknowledgments
The authors are thankful to M. Gruebe, S. Freist, S. Driesslein, and J. Perovic for their support during the experiments.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Preliminary Protocol for Cryopreservation Used for First Experiments with Le. gibba 7742
Appendix A.1. Donor Culture Maintenance
As described in the Material and Methods section, Section 3.1.
Appendix A.2. Pre-Culture
A portion of the donor duckweed culture were transferred onto a liquid SH nutrition medium supplemented with 100 g/L sucrose, pH 5.7–5.8, and aseptically incubated for 16–18 h (overnight) in a climate chamber set with a 12/12 h diurnal light/dark cycle, a PPFD of approximately 50–60 μmol·m−2·s−1 with the spectrum provided in Figure S13, and a 26 °C/25 °C temperature for the light/dark periods.
Appendix A.3. PVS3 Preparation
To prepare 100 mL of PVS3, 50 g of sucrose was intensively mixed with 28 mL of the SH medium (pH 7.1–7.2) until sucrose was almost completely dissolved; 50 g of glycerol was added, and the resulting mixture was heated in a water bath at 90–95 °C with periodic shaking until complete sucrose dissolution. The clear solution was sterilized by autoclaving at 121 °C for 20 min and stored at 4 °C until usage.
Appendix A.4. Dehydration and Cooling Procedure
- Prepare foil strips (≈7 mm width, ≈70 mm length). Insert the foil strips (with tweezers) to cryo-tubes (2.0 mL) to obtain a “U-form” shape formed by the strip inside the cryo-tubes. Press the side parts of the “U-shaped” foil strip to the sides of the cryo-tube with tweezers (see graphic instruction on Figure S1). Transfer the pre-formed “U-shaped” foil insert to the container for autoclavation.
- Sterilize the prepared “U-shaped” foil insert by autoclavation.
- In the laminar cabinet, insert the autoclaved “U-shaped” foil insert into the sterile cryo-tube using sterile tweezers.
- In the laminar cabinet, transfer a portion of the pre-cultivated duckweed fronds to sterile filter paper using sterile tweezers for brief blotting (for 1–2 min).
- Transfer a portion of the blotted duckweed fronds (25–50 fronds) to the cryo-tube, supplemented with the “U-shaped” foil insert by inserting the fronds between “U-shape” parts of the foil insert close to the bottom of the cryo-tube.
- Clamp the edges of the foil strip at the top of the cryo-tube with sterile tweezers to form the foil pack with the fronds inside the cryo-tube (see graphic instruction in Figure S2).
- Add a portion of the 1 mL of PVS3 to the cryo-tube containing duckweed fronds packed in a foil insert.
- Load the opened test tube into the vacuum chamber (exicator after aseptic treatment, placed in the laminar cabinet).
- Apply a vacuum (as deep as possible using a common laboratory vacuum pump).
- Incubate the test tube under vacuum for 15 min at room temperature. Release the vacuum.
- Transfer the cryo-tube from the exicator, and cover the cryo-tube with the sterile cap.
- Incubate for 1 h and 45 min at room temperature.
- Transfer the cryo-tube with the duckweed to liquid nitrogen. Store the sample in liquid nitrogen for at least several days.
Appendix A.5. Rewarming, Washing, and Unloading
- Prepare a glass test tube (sterile, 50 mL, with foil cap) filled with 10 mL of a sterile solution of 1.2 M sucrose (washing solution).
- Prepare the sterile 9 g/L glucose solution, buffered by 1 g/L MES, pH 7.1–7.2 (by KOH).
- Remove the cryo-tube from the liquid nitrogen, and defrost by heating in a water bath set at 40 °C, keeping the tube submersed in the bath until the ice in the tube almost completely melts (visual observation).
- Immediately after defrosting (almost complete ice melting), briefly treat the cryo-tube with the antiseptic solution, open the cryo-tube in the laminar cabinet, and slightly open the foil pack in the cryo-tube with sterile tweezers by separating the edges of the foil strip.
- Immediately transfer the foil pack with duckweed from the cryo-tube to the glass test tube, fill with the 1.2 M sucrose solution (room temperature), and close the test tube using the cap.
- Incubate for 1 h at room temperature. During this time, most of the duckweed fronds should have separated from the foil insert and should start floating on the surface of the washing solution.
- In the laminar cabinet, remove the solution of sucrose from the test tube (leaving the duckweed fronds in the test tube) and add 10 mL of the 9 g/L glucose solution, buffered by 1 g/L MES (pH 7.1–7.2, room temperature) to the test tube.
- Transfer the test tube to the climate chamber, set at +25 ± 1 °C, in a light-protecting box for further unloading from the rest of the components of the PVS3. Incubate for 1 day in darkness.
- In the laminar cabinet, remove the solution of glucose from the test tube (leaving the duckweed fronds in the test tube), and add a fresh portion of 10 mL of the 9 g/L glucose solution (pH 7.1–7.2, room temperature).
- Transfer the test tube to the climate chamber, set at +25 ± 1 °C, in a light-protecting box for further unloading and revitalizing. Incubate for 3 days in darkness.
Appendix A.6. Early Regrowth
- In the laminar cabinet, remove the solution of glucose from the test tube with duckweed and add 10 mL of the liquid SH nutrition medium supplemented with 5 g/L sucrose and 5 g/L glucose (pH 7.1–7.2, room temperature).
- Transfer the test tubes to the climate chamber, set at 12/12 h diurnal light/dark cycle, PPFD approximately 50–60 μmol·m−2·s−1 with the spectrum provided in Figure S13, and 26 °C/25 °C temperature for the light/dark periods. Incubate for further observations.
Appendix B. Optimized Protocol for Cryopreservation of Lemnaceae Representatives
Appendix B.1. Donor Culture Maintenance
As described in the Material and Methods section, Section 3.1. The duration of the last subculture of the donor duckweed plants should be in the range of 1–2 weeks prior to transferring to the pre-cultivation stage.
Appendix B.2. Pre-Culture
A portion of the donor duckweed culture was transferred onto the liquid SH nutrition medium supplemented with 0.4 M sucrose and 5% glycerol (v/v), pH 5.7–5.8, and aseptically incubated for 36–48 h in a climate chamber, set at 12/12 h diurnal light/dark cycle, PPFD approximately 50–60 μmol·m−2·s−1 with the spectrum provided in Figure S13, and 26 °C/25 °C temperature for the light/dark periods.
Appendix B.3. PVS3 Preparation
To prepare 100 mL of PVS3, 50 g of sucrose was intensively mixed with 28 mL of the SH medium (pH 7.1–7.2) until the sucrose was almost completely dissolved; 50 g of glycerol was added and the resulting mixture was heated in a water bath at 90–95 °C with periodic shaking until complete sucrose dissolution. The clear solution was sterilized by autoclaving at 121 °C for 20 min and stored at 4 °C until usage.
Appendix B.4. Dehydration and Cooling Procedure
- Prepare the foil strips (≈7 mm width, ≈70 mm length). Make 15–20 perforations in the central part of the foil strip using a needle or the sharp and thin end if small tweezers (to accelerate and simplify this operation, we made a special many-needled tool for this purpose from a comb for animals). Insert the foil strips into the plastic centrifugal test tube (2.0 mL) to obtain the “U-shape” form of the strip inside the test tube. Insert a magnetic stirrer bar (7 mm) or another stick with the same diameter and U-shaped end into the test tube to press the foil strip onto the internal surface of the centrifugal test tube (see graphic instruction on Figure S6). Transfer the pre-formed foil insert to the container for autoclavation.
- Sterilize the prepared foil inserts by autoclavation.
- Sterilize swabs by autoclavation.
- In the laminar cabinet, transfer 1 mL of sterile PVS3 solution to the sterile cryo-tubes (2 mL). Close, and precool in ice.
- In the laminar cabinet, insert the autoclaved swabs into the sterile plastic centrifugal test tubes (2 mL) using sterile tweezers. Push the swabs down to the bottom of the centrifugal test tubes.
- In the laminar cabinet, transfer the portions of duckweed fronds from donor cultures (after the pre-cultivation stage) to the sterile centrifugal test tubes, supplemented with cotton wool, using autoclaved foil inserts (as single-use sampler) and sterile tweezers (see graphic instruction on Figure 2).
- Clamp the edges of the foil strips at the top of the centrifugal test tubes with sterile tweezers to form the foil packs with the fronds inside the centrifugal test tubes.
- Close the test tubes, and spin-dry the duckweed fronds by centrifugation for 3 min at 800× g at room temperature.
- In the laminar cabinet, transfer the foil packs with spin-dried duckweed fronds to the precooled-in-ice cryo-tubes filled with 1 mL of PVS3.
- Immediately close the cryo-tubes, and transfer it on ice in darkness.
- Incubate for 4 h on ice in darkness.
- Transfer the cryo-tubes with the duckweed to liquid nitrogen. Perform the operation under weak daylight (or dusk) or weak artificial light. Store the sample in liquid nitrogen for at least several days.
Appendix B.5. Rewarming, Washing, and Unloading
- Prepare a glass test tube (sterile, 50 mL, with foil cap) filled with 10 mL of a sterile 1.2 M sucrose solution (washing solution). Precool and keep on ice.
- Prepare the sterile 9 g/L glucose solution, buffered by 1 g/L MES, pH 7.1–7.2 (by KOH). Precool at ≈10 °C.
- Remove the cryo-tube from liquid nitrogen, and immediately defrost by heating in a water bath set at 40 °C, keeping the tube submersed in the bath until some signs of viscous fluidity of PVS3 in the cryo-tube appears (visual observation). Perform the operations under weak daylight (or dusk) or weak artificial light.
- Immediately after defrosting, briefly treat the cryo-tube with the antiseptic solution, open the cryo-tube in the laminar cabinet, and slightly open the foil pack in the cryo-tube with sterile tweezers by separating the edges of the foil strip. Perform the operations under weak daylight (or dusk) or weak artificial light.
- Immediately transfer the foil pack with duckweed from the cryo-tube to the precooled-in-ice glass test tube, fill with the 1.2 M sucrose solution, and close the test tube using the cap. Perform the operations under weak daylight (or dusk) or weak artificial light.
- Immediately transfer the glass test tube with rewarmed duckweed on ice in darkness.
- Incubate on ice in darkness for 1 h. During this time, most of the duckweed fronds should have separated from the surface of the foil pack and should start floating on the surface of the washing solution.
- In the laminar cabinet, remove the solution of sucrose from the test tube (leaving the duckweed fronds in the test tube), and add 10 mL of the 9 g/L glucose solution, buffered by 1g/L MES (pH 7.1–7.2, precooled at ≈10 °C) to the glass test tube with rewarmed duckweed. Perform the operations under weak daylight (or dusk) or weak artificial light.
- Transfer the test tube to the climate chamber, set at +25 ± 1 °C, in a light-protecting box for further unloading from the rest of the components of the PVS3. Incubate for 1 day in darkness.
- In the laminar cabinet, remove the solution of glucose from the test tube (leaving the duckweed fronds in the test tube), and add a fresh portion of 10 mL of the 9 g/L glucose solution, buffered by 1 g/L MES (pH 7.1–7.2, room temperature). Perform the operations under weak daylight (or dusk) or weak artificial light.
- Transfer the test tube to the climate chamber, set at +25 ± 1 °C, in a light-protecting box for further unloading and revitalizing. Incubate for 3 days in darkness.
Appendix B.6. Early Regrowth
- In the laminar cabinet, remove the solution of glucose from the test tube with duckweed and add 10 mL of the liquid SH nutrition medium supplemented with 5 g/L sucrose and 5 g/L glucose (pH 7.1–7.2). For Wolffia australiana 8730, the liquid SH medium, supplemented with 5 g/L sucrose, 5 g/L glucose, 0.5 g/L casein hydrolysate, and 0.5 g/L yeast extract (pH 7.1–7.2), should be added. Perform the operations under weak daylight (or dusk) or weak artificial light.
- Transfer the test tubes to the climate chamber, set at +29 ± 1 °C, for re-growth. Incubate in accordance with the pulsed illumination regime (see Figure 1) with the light spectrum provided in Figure S13 and at a temperature of +29 ± 1 °C for 6 days.
- Transfer the test tubes to the climate chamber, set at a 12/12 h diurnal light/dark cycle, PPFD approximately 50–60 μmol·m−2·s−1 with the spectrum provided in Figure S13, and 26 °C/25 °C temperature for the light/dark periods for further re-growth.
References
- Tippery, N.P.; Les, D.H. Tiny Plants with Enormous Potential: Phylogeny and Evolution of Duckweeds. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer: Cham, Switzerland, 2020; pp. 19–38. ISBN 978-3-030-11044-4. [Google Scholar]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.K.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. Plant Cell 2021, 33, 3207–3234. [Google Scholar] [CrossRef]
- Zhou, Y.; Stepanenko, A.; Kishchenko, O.; Xu, J.; Borisjuk, N. Duckweeds for Phytoremediation of Polluted Water. Plants 2023, 12, 589. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.; Kishchenko, O.; Zhou, Y.; Vasylenko, M.; Giritch, A.; Sun, J.; Borisjuk, N.; Kuchuk, M. Robust Agrobacterium-Mediated Transient Expression in Two Duckweed Species (Lemnaceae) Directed by Non-replicating, Replicating, and Cell-to-Cell Spreading Vectors. Front. Bioeng. Biotechnol. 2021, 9, 761073. [Google Scholar] [CrossRef] [PubMed]
- Braglia, L.; Breviario, D.; Gianì, S.; Gavazzi, F.; De Gregori, J.; Morello, L. New Insights into Interspecific Hybridization in Lemna L. Sect. Lemna (Lemnaceae Martinov). Plants 2021, 10, 2767. [Google Scholar] [CrossRef] [PubMed]
- Braglia, L.; Lauria, M.; Appenroth, K.J.; Bog, M.; Breviario, D.; Grasso, A.; Gavazzi, F.; Morello, L. Duckweed Species Genotyping and Interspecific Hybrid Discovery by Tubulin-Based Polymorphism Fingerprinting. Front. Plant Sci. 2021, 12, 625670. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Appenroth, K.J.; Ma, Y.; Shoham, T.; Sree, K.S. Registration of Duckweed Clones/Strains—Future Approach. Duckweed Forum 2020, 8, 35–37. [Google Scholar]
- Stomp, A.M. The duckweeds: A valuable plant for biomanufacturing. In Biotechnology Annual Review; Elsevier: Amsterdam, The Netherlands, 2005; Volume 11, pp. 69–99. [Google Scholar]
- Yang, G.-L.; Feng, D.; Liu, Y.-T.; Lv, S.-M.; Zheng, M.-M.; Tan, A.-J. Research Progress of a Potential Bioreactor: Duckweed. Biomolecules 2021, 11, 93. [Google Scholar] [CrossRef]
- Yang, J.; Hu, S.; Li, G.; Khan, S.; Kumar, S.; Yao, L.; Duan, P.; Hou, H. Transformation Development in Duckweeds. In The Duckweed Genomes; Cao, X.H., Fourounjian, P., Wang, W., Eds.; Springer: Cham, Switzerland, 2020; pp. 143–155. ISBN 978-3-030-11044-4. [Google Scholar]
- Yu, L.; Wang, Y.; Xu, S.; Tang, X.; Zhao, J.; Yu, C.; He, G.; Xu, H.; Wang, S.; Tang, Y.; et al. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing in Lemna aequinoctialis. Plant Biotechnol. J. 2019, 17, 2143–2152. [Google Scholar] [CrossRef]
- Shahak, Y.; Posner, H.B.; Avron, M. Evidence for a Block between Plastoquinone and Cytochrome f in a Photosynthetic Mutant of Lemna with Abnormal Flowering Behavior. Plant Physiol. 1976, 57, 577–579. [Google Scholar] [CrossRef][Green Version]
- Tam, Y.Y.; Slovin, J.P.; Cohen, J.D. Selection and Characterization of [alpha]-Methyltryptophan-Resistant Lines of Lemna gibba Showing a Rapid Rate of Indole-3-Acetic Acid Turnover. Plant Physiol. 1995, 107, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Panis, B.; Nagel, M.; Van den Houwe, I. Challenges and Prospects for the Conservation of Crop Genetic Resources in Field Genebanks, in In Vitro Collections and/or in Liquid Nitrogen. Plants 2020, 9, 1634. [Google Scholar] [CrossRef]
- Reed, B.M. (Ed.) Plant Cryopreservation: A Practical Guide; Springer: New York, NY, USA, 2008; ISBN 978-0-387-72275-7. [Google Scholar]
- Volk, G.M.; Walters, C. Plant vitrification solution 2 lowers water content and alters freezing behavior in shoot tips during cryoprotection. Cryobiology 2006, 52, 48–61. [Google Scholar] [CrossRef]
- Kreckel, H.D.; Samuels, F.M.D.; Bonnart, R.; Volk, G.M.; Stich, D.G.; Levinger, N.E. Tracking Permeation of Dimethyl Sulfoxide (DMSO) in Mentha × piperita Shoot Tips Using Coherent Raman Microscopy. Plants 2023, 12, 2247. [Google Scholar] [CrossRef]
- Tunçer, S.; Gurbanov, R.; Sheraj, I.; Solel, E.; Esenturk, O.; Banerjee, S. Low dose dimethyl sulfoxide driven gross molecular changes have the potential to interfere with various cellular processes. Sci. Rep. 2018, 8, 14828. [Google Scholar] [CrossRef] [PubMed]
- Zamecnik, J.; Faltus, M.; Bilavcik, A. Vitrification Solutions for Plant Cryopreservation: Modification and Properties. Plants 2021, 10, 2623. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, S.; Sakai, A.; Amano, Y.; Matsuzawa, T. Cryopreservation of asparagus (Asparagus officinalis L.) embryogenic suspension cells and subsequent plant regeneration by vitrification. Plant Sci. 1993, 91, 67–73. [Google Scholar] [CrossRef]
- Borisjuk, N.; Peterson, A.A.; Lv, J.; Qu, G.; Luo, Q.; Shi, L.; Chen, G.; Kishchenko, O.; Zhou, Y.; Shi, J. Structural and Biochemical Properties of Duckweed Surface Cuticle. Front. Chem. 2018, 6, 317. [Google Scholar] [CrossRef]
- Sauter, P.R. Kryokonservierung von Lemnaceae. Ph.D. Thesis, ETH Zürich, Zürich, Switzerland, 1993. [Google Scholar] [CrossRef]
- Parsons, J.; Wingate, V. Methods and Compositions for the Cryopreservation of Duckweed 2011. Patent No. WO2011005502A3, 29 December 2011. [Google Scholar]
- Ito, S.; Tanaka, D.; Oyama, T. Development of Cryopreservation Protocol for a Variety of Duckweed Meristems by Vitrification-cryo-Plate Method. In Proceedings of the 6th International Conference on Duckweed Research and Application ICDRA 2022, Gatersleben, Germany, 29 May–1 June 2022; p. 34. [Google Scholar]
- Nadarajan, J.; Pritchard, H.W. Biophysical Characteristics of Successful Oilseed Embryo Cryoprotection and Cryopreservation Using Vacuum Infiltration Vitrification: An Innovation in Plant Cell Preservation. PLoS ONE 2014, 9, e96169. [Google Scholar] [CrossRef] [PubMed]
- Volk, G. Application of Functional Genomics and Proteomics to Plant Cryopreservation. Curr. Genom. 2010, 11, 24–29. [Google Scholar] [CrossRef]
- Bruňáková, K.; Čellárová, E. Conservation Strategies in the Genus Hypericum via Cryogenic Treatment. Front. Plant Sci. 2016, 7, 558. [Google Scholar] [CrossRef]
- Roque-Borda, C.A.; Kulus, D.; de Souza, A.V.; Kaviani, B.; Vicente, E.F. Cryopreservation of Agronomic Plant Germplasm Using Vitrification-Based Methods: An Overview of Selected Case Studies. Int. J. Mol. Sci. 2021, 22, 6157. [Google Scholar] [CrossRef] [PubMed]
- Barrero-Gil, J.; Salinas, J. Gene Regulatory Networks Mediating Cold Acclimation: The CBF Pathway. In Survival Strategies in Extreme Cold and Desiccation; Iwaya-Inoue, M., Sakurai, M., Uemura, M., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; Volume 1081, pp. 3–22. ISBN 978-9-81131-243-4. [Google Scholar]
- Vyse, K.; Schaarschmidt, S.; Erban, A.; Kopka, J.; Zuther, E. Specific CBF transcription factors and cold-responsive genes fine-tune the early triggering response after acquisition of cold priming and memory. Physiol. Plant. 2022, 174, e13740. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-Induced CBF–PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, Q.; Hao, J.; Zhu, X.; Cheng, Y.; Mao, J.; Lin, M.; Xia, E.; Li, Y. Genome-Wide Characterization of the C-repeat Binding Factor (CBF) Gene Family Involved in the Response to Abiotic Stresses in Tea Plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBF s in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Baker, C.R.; Stewart, J.J.; Amstutz, C.L.; Ching, L.G.; Johnson, J.D.; Niyogi, K.K.; Adams, W.W.; Demmig-Adams, B. Genotype-dependent contribution of CBF transcription factors to long-term acclimation to high light and cool temperature. Plant Cell Environ. 2022, 45, 392–411. [Google Scholar] [CrossRef] [PubMed]
- Lemon, G.D.; Posluszny, U. Comparative Shoot Development and Evolution in the Lemnaceae. Int. J. Plant Sci. 2000, 161, 733–748. [Google Scholar] [CrossRef]
- Ren, L.; Wang, M.-R.; Wang, Q.-C. ROS-induced oxidative stress in plant cryopreservation: Occurrence and alleviation. Planta 2021, 254, 124. [Google Scholar] [CrossRef]
- Volk, G.M.; Harris, J.L.; Rotindo, K.E. Survival of mint shoot tips after exposure to cryoprotectant solution components. Cryobiology 2006, 52, 305–308. [Google Scholar] [CrossRef]
- Senula, A.; Nagel, M. Cryopreservation of Plant Shoot Tips of Potato, Mint, Garlic, and Shallot Using Plant Vitrification Solution 3. In Cryopreservation and Freeze-Drying Protocols; Wolkers, W.F., Oldenhof, H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2180, pp. 647–661. ISBN 978-1-07-160782-4. [Google Scholar]
- Lukić, M.; Funnekotter, B.; Considine, M.J.; Bunn, E.; Mancera, R.L. Evaluation of Vacuum Infiltration Vitrification and Cryo-mesh Cryopreservation Techniques with Arabidopsis thaliana Shoot Tips. Cryo Lett. 2022, 43, 328–333. [Google Scholar] [CrossRef]
- Normah, M.N.; Sulong, N.; Reed, B.M. Cryopreservation of shoot tips of recalcitrant and tropical species: Advances and strategies. Cryobiology 2019, 87, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, R.A.; Kalisz-Nowak, B.; Korohoda, W.; Wieckowski, S.; Kilarski, W.; Kozlowska, M. Contractile elements of Lemna trisulca L. glycerinated cell models during chloroplast translocations. Biochem. Cell Biol. 2000, 78, 503–510. [Google Scholar] [CrossRef]
- Yamada, T.; Marubashi, W.; Niwa, M. Detection of Four Lethality Types in Interspecific Crosses among Nicotiana Species through the Use of Three Rescue Methods for Lethality. Breed. Sci. 1999, 49, 203–210. [Google Scholar] [CrossRef]
- Folgado, R.; Panis, B.; Sergeant, K.; Renaut, J.; Swennen, R.; Hausman, J.-F. Unravelling the effect of sucrose and cold pretreatment on cryopreservation of potato through sugar analysis and proteomics. Cryobiology 2015, 71, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakière, B.; Vanacker, H.; Miginiac-Maslow, M.; Van Breusegem, F.; Noctor, G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cel: Photoperiod and H2O2 signalling. Plant J. 2007, 52, 640–657. [Google Scholar] [CrossRef]
- Challabathula, D.; Puthur, J.T.; Bartels, D. Surviving metabolic arrest: Photosynthesis during desiccation and rehydration in resurrection plants: Surviving Metabolic Arrest. Ann. N. Y. Acad. Sci. 2016, 1365, 89–99. [Google Scholar] [CrossRef]
- Rogers, H.; Munné-Bosch, S. Production and Scavenging of Reactive Oxygen Species and Redox Signaling during Leaf and Flower Senescence: Similar but Different. Plant Physiol. 2016, 171, 1560–1568. [Google Scholar] [CrossRef]
- Mechela, A.; Schwenkert, S.; Soll, J. A brief history of thylakoid biogenesis. Open Biol. 2019, 9, 180237. [Google Scholar] [CrossRef]
- Köpnick, C.; Grübe, M.; Stock, J.; Senula, A.; Mock, H.-P.; Nagel, M. Changes of soluble sugars and ATP content during DMSO droplet freezing and PVS3 droplet vitrification of potato shoot tips. Cryobiology 2018, 85, 79–86. [Google Scholar] [CrossRef]
- Madsen, J.D. Biomass Techniques for Monitoring and Assessing Control of Aquatic Vegetation. Lake Reserv. Manag. 1993, 7, 141–154. [Google Scholar] [CrossRef]
- Turner, S.; Senaratna, T.; Touchell, D.; Bunn, E.; Dixon, K.; Tan, B. Stereochemical arrangement of hydroxyl groups in sugar and polyalcohol molecules as an important factor in effective cryopreservation. Plant Sci. 2001, 160, 489–497. [Google Scholar] [CrossRef]
- Sarin, L.P.; Sree, K.S.; Bóka, K.; Keresztes, Á.; Fuchs, J.; Tyagi, A.K.; Khurana, J.P.; Appenroth, K.-J. Characterisation of a Spontaneous Mutant of Lemna gibba G3 (Lemnaceae). Plants 2023, 12, 2525. [Google Scholar] [CrossRef]
- Ernst, E.; Abramson, B.; Acosta, K.; Hoang, P.T.N.; Mateo-Elizalde, C.; Schubert, V.; Pasaribu, B.; Hartwick, N.; Colt, K.; Aylward, A.; et al. The Genomes and Epigenomes of Aquatic Plants (Lemnaceae) Promote Triploid Hybridization and Clonal Repro-duction. Plant Biol. 2023, 2023, 551673. [Google Scholar] [CrossRef]
- Sror, H.A.M.; Tischendorf, G.; Sieg, F.; Schmitt, J.M.; Hincha, D.K. Cryoprotectin protects thylakoids during a freeze–thaw cycle by a mechanism involving stable membrane binding. Cryobiology 2003, 47, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Sershen; Berjak, P.; Pammenter, N.W.; Wesley-Smith, J. The effects of various parameters during processing for cryopreservation on the ultrastructure and viability of recalcitrant zygotic embryos of Amaryllis belladonna. Protoplasma 2012, 249, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Heringer, A.S.; Steinmacher, D.A.; Schmidt, É.C.; Bouzon, Z.L.; Guerra, M.P. Survival and ultrastructural features of peach palm (Bactris gasipaes, Kunth) somatic embryos submitted to cryopreservation through vitrification. Protoplasma 2013, 250, 1185–1193. [Google Scholar] [CrossRef]
- Stock, J.; Bräutigam, A.; Melzer, M.; Bienert, G.P.; Bunk, B.; Nagel, M.; Overmann, J.; Keller, E.R.J.; Mock, H.-P. The transcription factor WRKY22 is required during cryo-stress acclimation in Arabidopsis shoot tips. J. Exp. Bot. 2020, 71, 4993–5009. [Google Scholar] [CrossRef] [PubMed]
- Ingle, R.A.; Collett, H.; Cooper, K.; Takahashi, Y.; Farrant, J.M.; Illing, N. Chloroplast biogenesis during rehydration of the resurrection plant Xerophyta humilis: Parallels to the etioplast-chloroplast transition. Plant Cell Environ. 2008, 31, 1813–1824. [Google Scholar] [CrossRef]
- Charuvi, D.; Kiss, V.; Nevo, R.; Shimoni, E.; Adam, Z.; Reich, Z. Gain and Loss of Photosynthetic Membranes during Plastid Differentiation in the Shoot Apex of Arabidopsis. Plant Cell 2012, 24, 1143–1157. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, M.-R.; Li, J.; Cui, Z.; Volk, G.M.; Wang, Q. Cryobiotechnology: A Double-Edged Sword for Obligate Plant Pathogens. Plant Dis. 2019, 103, 1058–1067. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Kidokoro, S.; Konoura, I.; Soma, F.; Suzuki, T.; Miyakawa, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in Arabidopsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2216183120. [Google Scholar] [CrossRef] [PubMed]
- Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The Influence of Light Quality, Circadian Rhythm, and Photoperiod on the CBF-Mediated Freezing Tolerance. Int. J. Mol. Sci. 2013, 14, 11527–11543. [Google Scholar] [CrossRef]
- Lee, E.S.; Park, J.H.; Wi, S.D.; Kang, C.H.; Chi, Y.H.; Chae, H.B.; Paeng, S.K.; Ji, M.G.; Kim, W.-Y.; Kim, M.G.; et al. Redox-dependent structural switch and CBF activation confer freezing tolerance in plants. Nat. Plants 2021, 7, 914–922. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef]
- Doležel, J.; Bartoš, J.; Voglmayr, H.; Greilhuber, J. Letter to the editor. Cytometry 2003, 51, 127–128. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Baker, N.R. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef]
- Murchie, E.H.; Lawson, T. Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef] [PubMed]
- Tschiersch, H.; Junker, A.; Meyer, R.C.; Altmann, T. Establishment of integrated protocols for automated high throughput kinetic chlorophyll fluorescence analyses. Plant Methods 2017, 13, 54. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.; Lyons, E.; Bomhoff, M.; Lenards, A. Comparative Genomics of Grass Genomes Using CoGe. In Handbook of Plant and Crop Physiology; CRC Press: Boca Raton, FL, USA, 2014; pp. 797–816. ISBN 978-0-429-09788-1. [Google Scholar]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- GenomeNet. Available online: https://www.genome.jp/tools/ete/ (accessed on 1 May 2022).
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45e–445e. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).