OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance
Abstract
:1. Introduction
2. Results
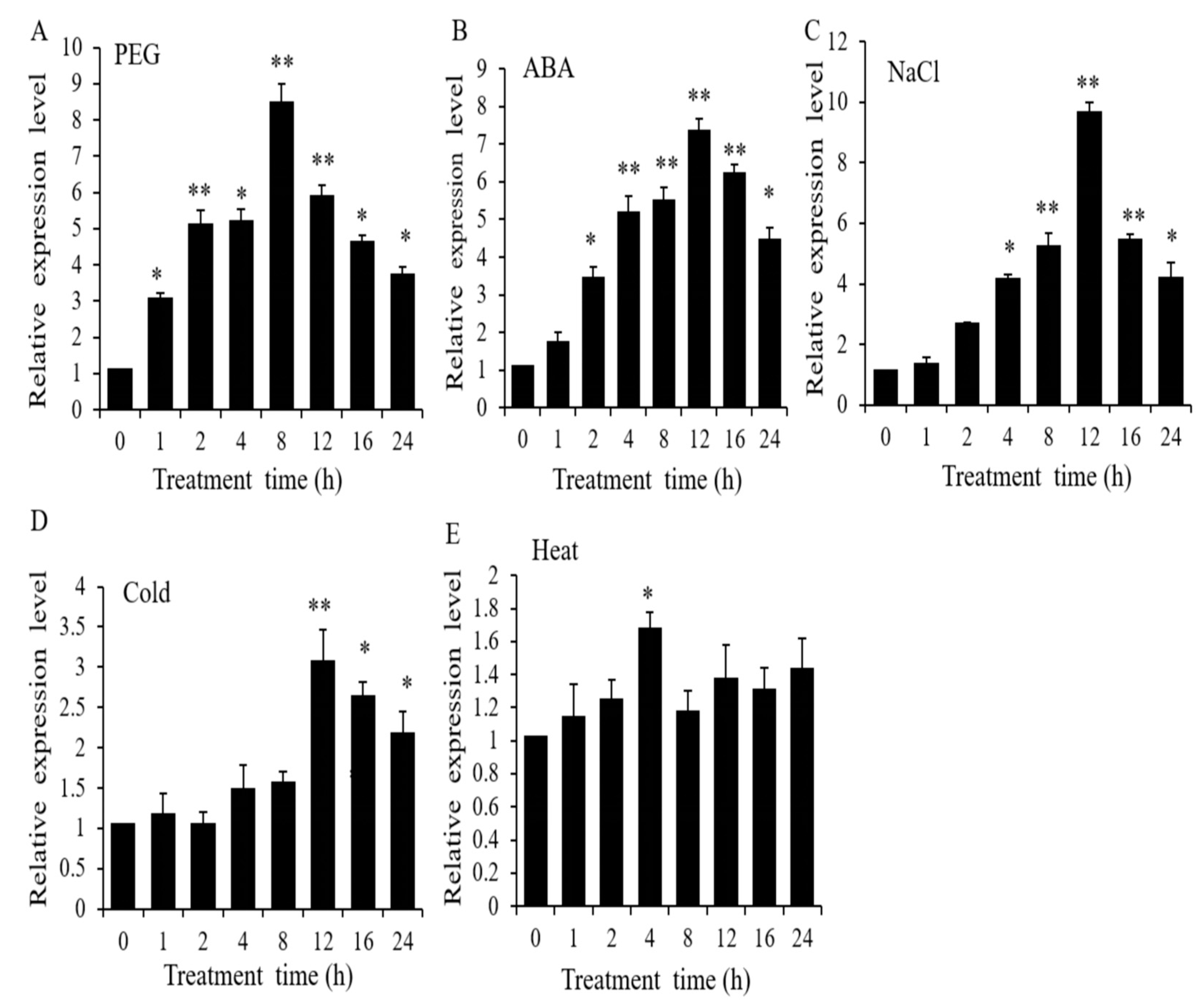
2.1. Expression Patterns of OsWRKY97
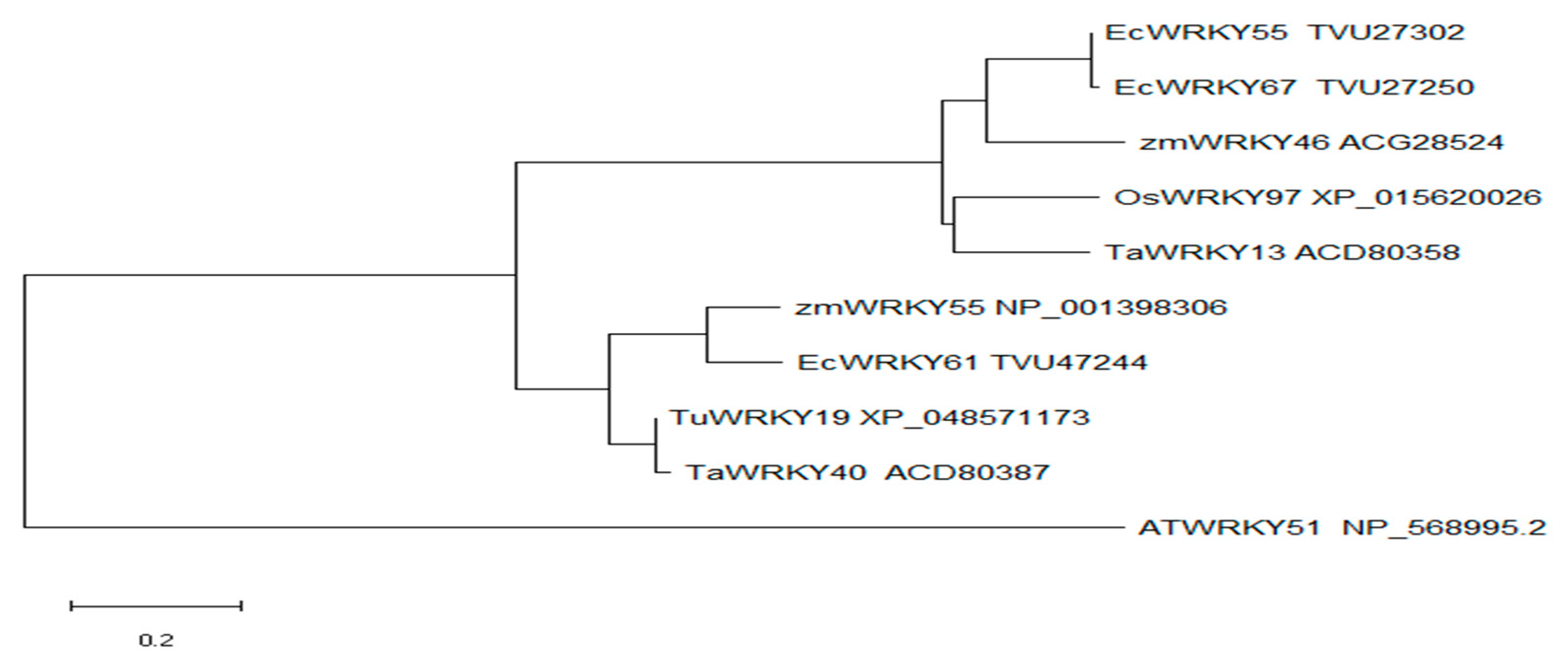
2.2. Phylogenetic Analysis of OsWRKY97
2.3. Subcellular Localization of OsWRKY97
2.4. Analysis of OsWRKY97 Promoter Domain
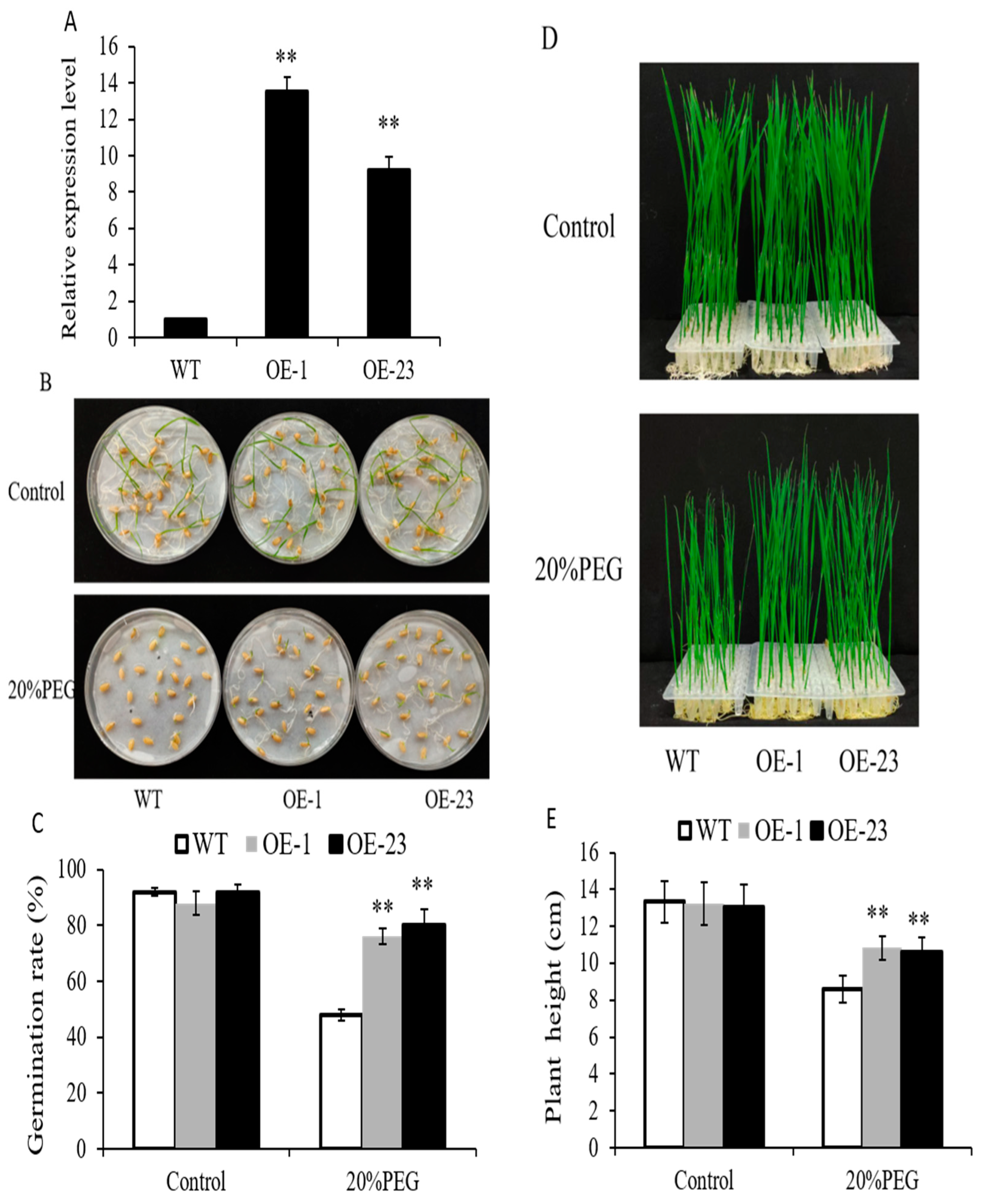
2.5. Overexpression of OsWRKY97 in Rice Improved Osmotic Stress Tolerance at the Germination and Post-Germination Stages
2.6. Overexpression of OsWRKY97 in Rice Enhanced Drought Stress Tolerance
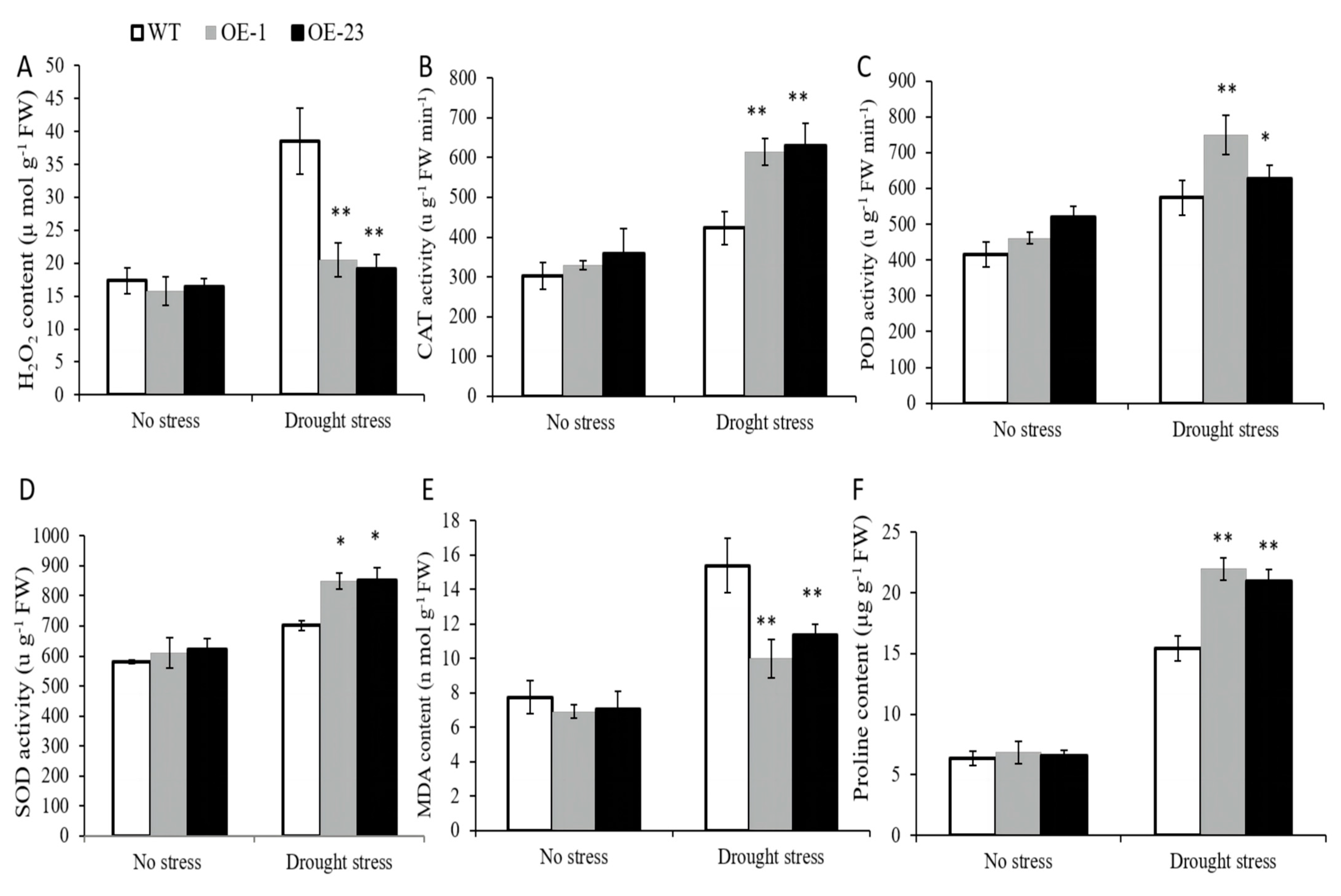
2.7. Effects of OsWRKY97 Overexpression on Related Physiological Indexes under Drought Stress
2.8. OsWRKY97 Is a Positive Regulator in ABA Signaling under Drought Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Abiotic Treatments
4.3. RNA Extraction and Real-Time PCR
4.4. Sequence Analysis of OsWRKY97
4.5. Vector Construction and Gene Transformation
4.6. Determination of Stress-Associated Physiological Indicators
4.7. Measurement of ABA Content
4.8. Subcellular Localization of OsWRKY97
4.9. Analysis of Stress Tolerance
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic Mechanisms of Abiotic Stress Tolerance That Translate to Crop Yield Stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiong, L. General Mechanisms of Drought Response and Their Application in Drought Resistance Improvement in Plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling During Cold, Drought, and Salt Stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early Abscisic Acid Signal Transduction Mechanisms: Newly Discovered Components and Newly Emerging Questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef]
- Mizuno, T.; Yamashino, T. Comparative Transcriptome of Diurnally Oscillating Genes and Hormone-Responsive Genes in Arabidopsis Thaliana: Insight into Circadian Clock-Controlled Daily Responses to Common Ambient Stresses in Plants. Plant Cell Physiol. 2008, 49, 481–487. [Google Scholar] [CrossRef]
- Desikan, R.; Cheung, M.K.; Bright, J.; Henson, D.; Hancock, J.T.; Neill, S.J. ABA, Hydrogen Peroxide and Nitric Oxide Signalling in Stomatal Guard Cells. J. Exp. Bot. 2004, 55, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, D.W.A.; Hills, A.; Köhler, B.; Blatt, M.R. Ca2+ Channels at the Plasma Membrane of Stomatal Guard Cells Are Activated by Hyperpolarization and Abscisic Acid. Proc. Natl. Acad. Sci. USA 2000, 97, 4967–4972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.-P. Hydrogen Peroxide Is Involved in Abscisic Acid-Induced Stomatal Closure in Vicia Faba. Plant. Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Wei, C.; Liu, X.; Wang, M.; Yu, F.; Xie, Q.; Tu, J. The Ring Finger Ubiquitin E3 Ligase Oshtas Enhances Heat Tolerance by Promoting H2O2-Induced Stomatal Closure in Rice. Plant Physiol. 2016, 170, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Sun, W.; Yang, G.; Sun, J. WRKY Transcription Factors in Legumes. BMC Plant Biol. 2018, 18, 243. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.-Q.; Chen, Z. Protein-Protein Interactions in the Regulation of WRKY Transcription Factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY Transcription Factor TaWRKY2 Enhances Drought Stress Tolerance in Transgenic Wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, M.A.; Song, A.; Faheem, M.; Chen, S.; Jiang, J.; Liu, C.; Fan, Q.; Chen, F. Involvement of CmWRKY10 in Drought Tolerance of Chrysanthemum through the ABA-Signaling Pathway. Int. J. Mol. Sci. 2016, 17, 693. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 Alleles Play Different Roles in Abscisic Acid Signalling and Salt Stress Tolerance but Similar Roles in Drought and Cold Tolerance in Rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 Are Master Transcription Factors That Cooperatively Regulate ABRE-Dependent ABA Signaling Involved in Drought Stress Tolerance and Require ABA for Full Activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA Signaling in Stress-Response and Seed Development. Plant Cell. Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.-K. Arabidopsis Mutant Deficient in 3 Abscisic Acid-Activated Protein Kinases Reveals Critical Roles in Growth, Reproduction, and Stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef]
- Antoni, R.; Rodriguez, L.; Gonzalez-Guzman, M.; Pizzio, G.A.; Rodriguez, P.L. News on ABA Transport, Protein Degradation and ABFs/WRKYs in ABA Signaling. Curr. Opin. Plant Biol. 2011, 14, 547–553. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.-K.; Gong, Z. ABO3, a WRKY Transcription Factor, Mediates Plant Responses to Abscisic Acid and Drought Tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.-T.; Liang, S.; Zhang, X.-F.; Wang, X.-F.; Zhang, D.-P. Cooperation of Three WRKY-Domain Transcription Factors WRKY18, WRKY40, and WRKY60 in Repressing Two ABA-Responsive Genes Abi4 and Abi5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef]
- Yao, L.; Cheng, X.; Gu, Z.; Huang, W.; Li, S.; Wang, L.; Wang, Y.-F.; Xu, P.; Ma, H.; Ge, X. The AWPM-19 Family Protein OsPM1 Mediates Abscisic Acid Influx and Drought Response in Rice. Plant Cell 2018, 30, 1258–1276. [Google Scholar] [CrossRef]
- Miller, G.A.D.; Mittler, R.O.N. Could Heat Shock Transcription Factors Function as Hydrogen Peroxide Sensors in Plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhou, L.; Liu, J.; Du, X.; Huang, F.; Pan, G.; Cheng, F. Relationship of ROS Accumulation and Superoxide Dismutase Isozymes in Developing Anther with Floret Fertility of Rice under Heat Stress. Plant Physiol. Biochem. 2018, 122, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-Cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Mundy, J.; Chua, N.H. Abscisic Acid and Water-Stress Induce the Expression of a Novel Rice Gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant Responses to Drought, Salinity and Extreme Temperatures: Towards Genetic Engineering for Stress Tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, H.J.; Gong, Q.; Li, P.; Ma, S. Unraveling Abiotic Stress Tolerance Mechanisms--Getting Genomics Going. Curr. Opin. Plant Biol. 2006, 9, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Jiang, W.; Yu, D. Male Gametophyte-Specific WRKY34 Transcription Factor Mediates Cold Sensitivity of Mature Pollen in Arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X.; Zhou, X.; Chu, C.; Wang, X. OsWRKY30 Is Activated by Map Kinases to Confer Drought Tolerance in Rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.-S.; Park, S.R.; Lee, S.; Hwang, D.-J. Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M.; et al. Expression of Wild Soybean WRKY20 in Arabidopsis Enhances Drought Tolerance and Regulates ABA Signalling. J. Exp. Bot. 2013, 64, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-Mediated Transcriptional Regulation in Response to Osmotic Stress in Plants. J. Plant. Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- León, J.; Castillo, M.C.; Coego, A.; Lozano-Juste, J.; Mir, R. Diverse Functional Interactions between Nitric Oxide and Abscisic Acid in Plant Development and Responses to Stress. J. Exp. Bot. 2014, 65, 907–921. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, N.; Liu, R.; Chen, M.; Zhang, J. H2O2 Mediates the Regulation of ABA Catabolism and GA Biosynthesis in Arabidopsis Seed Dormancy and Germination. J. Exp. Bot. 2010, 61, 2979–2990. [Google Scholar] [CrossRef] [PubMed]
- Saxena, I.; Srikanth, S.; Chen, Z. Cross Talk between H2O2 and Interacting Signal Molecules under Plant Stress Response. Front. Plant Sci. 2016, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. Nadph Oxidase Atrbohd and Atrbohf Genes Function in ROS-Dependent ABA Signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Ozfidan, C.; Turkan, I.; Sekmen, A.H.; Seckin, B. Abscisic Acid-Regulated Responses of Aba2-1 under Osmotic Stress: The Abscisic Acid-Inducible Antioxidant Defence System and Reactive Oxygen Species Production. Plant Biol. 2012, 14, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.; Ludidi, N. Drought and Exogenous Abscisic Acid Alter Hydrogen Peroxide Accumulation and Differentially Regulate the Expression of Two Maize Rd22-Like Genes. Sci. Rep. 2017, 7, 8821. [Google Scholar] [CrossRef] [PubMed]
- Kakar, K.U.; Ren, X.L.; Nawaz, Z.; Cui, Z.Q.; Li, B.; Xie, G.L.; Hassan, M.A.; Ali, E.; Sun, G.C. A Consortium of Rhizobacterial Strains and Biochemical Growth Elicitors Improve Cold and Drought Stress Tolerance in Rice (Oryza sativa L.). Plant Biol. 2016, 18, 471–483. [Google Scholar] [CrossRef]
- Yoshida, S. Laboratory Manual for Physiological Studies of Rice, 3rd ed.; International Rice Research Institute: Los Banos, Philippines, 1976; pp. 61–67. [Google Scholar]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-Expression of a Single Ca2+ -Dependent Protein Kinase Confers Both Cold and Salt/Drought Tolerance on Rice Plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y.; et al. A Maize Phytochrome-Interacting Factors Protein Zmpif1 Enhances Drought Tolerance by Inducing Stomatal Closure and Improves Grain Yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2−Δδct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ozawa, K. A High-Efficiency Agrobacterium-Mediated Transformation System of Rice (Oryza sativa L.). Methods Mol. Biol. 2012, 847, 51–57. [Google Scholar] [PubMed]
- Du, H.; Wang, N.; Cui, F.; Li, X.; Xiao, J.; Xiong, L. Characterization of the Beta-Carotene Hydroxylase Gene Dsm2 Conferring Drought and Oxidative Stress Resistance by Increasing Xanthophylls and Abscisic Acid Synthesis in Rice. Plant Physiol. 2010, 154, 1304–1318. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, Y.; Lin, C.; Pan, R.; Ma, W.; Zheng, Y.; Guan, Y.; Hu, J. Suppression of Lox Activity Enhanced Seed Vigour and Longevity of Tobacco (Nicotiana tabacum L.) Seeds During Storage. Conserv. Physiol. 2018, 6, coy047. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF Transcription Factor TaERF3 Promotes Tolerance to Salt and Drought Stresses in Wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef]
- García, M.N.M.; Cortelezzi, J.I.; Fumagalli, M.; Capiati, D.A. Expression of the Arabidopsis ABF4 Gene in Potato Increases Tuber Yield, Improves Tuber Quality and Enhances Salt and Drought Tolerance. Plant Mol. Biol. 2018, 98, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, G.; Xia, X.; Wang, M.; Yin, X.; Zhang, B.; Zhang, X.; Cui, Y. Deciphering the Physiological and Molecular Mechanisms for Copper Tolerance in Autotetraploid Arabidopsis. Plant Cell. Rep. 2017, 36, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- López-Carbonell, M.; Jáuregui, O. A Rapid Method for Analysis of Abscisic Acid (ABA) in Crude Extracts of Water Stressed Arabidopsis Thaliana Plants by Liquid Chromatography--Mass Spectrometry in Tandem Mode. Plant Physiol. Bioch. 2005, 43, 407–411. [Google Scholar] [CrossRef]
- He, X.; Li, L.; Xu, H.; Xi, J.; Cao, X.; Xu, H.; Rong, S.; Dong, Y.; Wang, C.; Chen, R.; et al. A Rice Jacalin-Related Mannose-Binding Lectin Gene, Osjrl, Enhances Escherichia Coli Viability under High Salinity Stress and Improves Salinity Tolerance of Rice. Plant Biol. 2017, 19, 257–267. [Google Scholar] [CrossRef] [PubMed]



| Elements | Sequence | Function |
|---|---|---|
| MYB | WAACCA/YAACKG/CTAACCA/CNGTTR/AACGG/TAACAAA/TAACAAA/MACCWAMC/CCWACC/GGATA | ABA and drought-responsive elements |
| CGTCA-motif | CGTCA | Involved the MeJA-responsiveness |
| MYC | CATGTG/CANNTG | ABA and drought-responsive elements |
| GARE | TAACAA | GA-responsive element |
| W-Box | TTTGACY/TTGAC/CTGACY/TGACY | SA-responsive element |
| TC-rich repeats | GTTTTCTTAC | involved in defense and stress-responsiveness |
| TCA-element | CCATCTTTTT | involved in SA-responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, M.; Hou, D.; Wan, J.; Ye, T.; Zhang, L.; Fan, J.; Li, C.; Dong, Y.; Chen, W.; Rong, S.; et al. OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants 2023, 12, 3338. https://doi.org/10.3390/plants12183338
Lv M, Hou D, Wan J, Ye T, Zhang L, Fan J, Li C, Dong Y, Chen W, Rong S, et al. OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants. 2023; 12(18):3338. https://doi.org/10.3390/plants12183338
Chicago/Turabian StyleLv, Miaomiao, Dejia Hou, Jiale Wan, Taozhi Ye, Lin Zhang, Jiangbo Fan, Chunliu Li, Yilun Dong, Wenqian Chen, Songhao Rong, and et al. 2023. "OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance" Plants 12, no. 18: 3338. https://doi.org/10.3390/plants12183338
APA StyleLv, M., Hou, D., Wan, J., Ye, T., Zhang, L., Fan, J., Li, C., Dong, Y., Chen, W., Rong, S., Sun, Y., Xu, J., Cai, L., Gao, X., Zhu, J., Huang, Z., Xu, Z., & Li, L. (2023). OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants, 12(18), 3338. https://doi.org/10.3390/plants12183338






