Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas
Abstract
:1. Introduction
2. Results
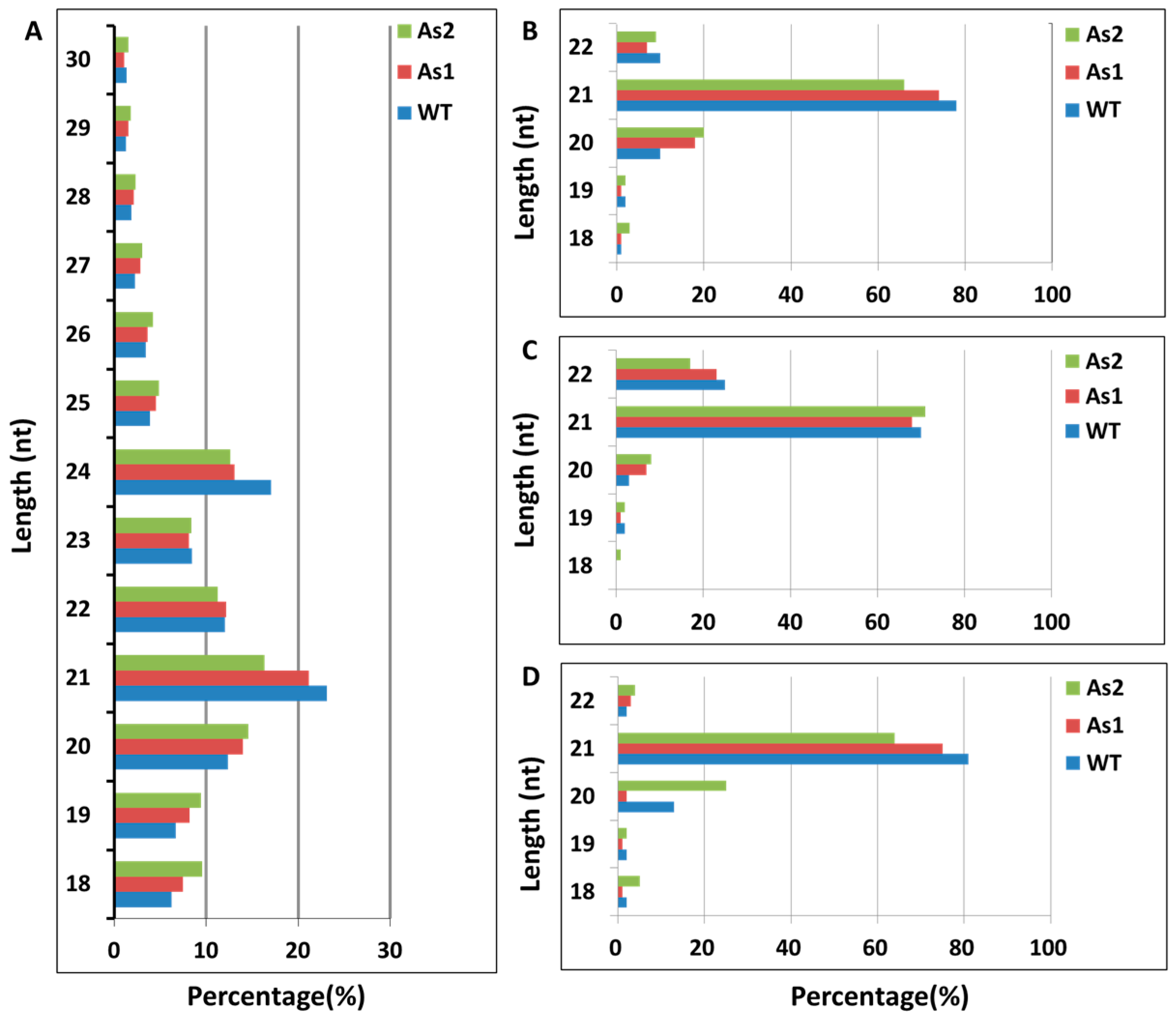
2.1. Acquisition of Total RNA and Distribution of Small RNA Fragment Length in Banana Fruit
2.2. Classification and Annotation of Small RNAs in Wild-Type and RNAi Transgenic Bananas
2.3. Classification and Annotation of Small RNAs in Wild-Type and RNAi Transgenic Bananas
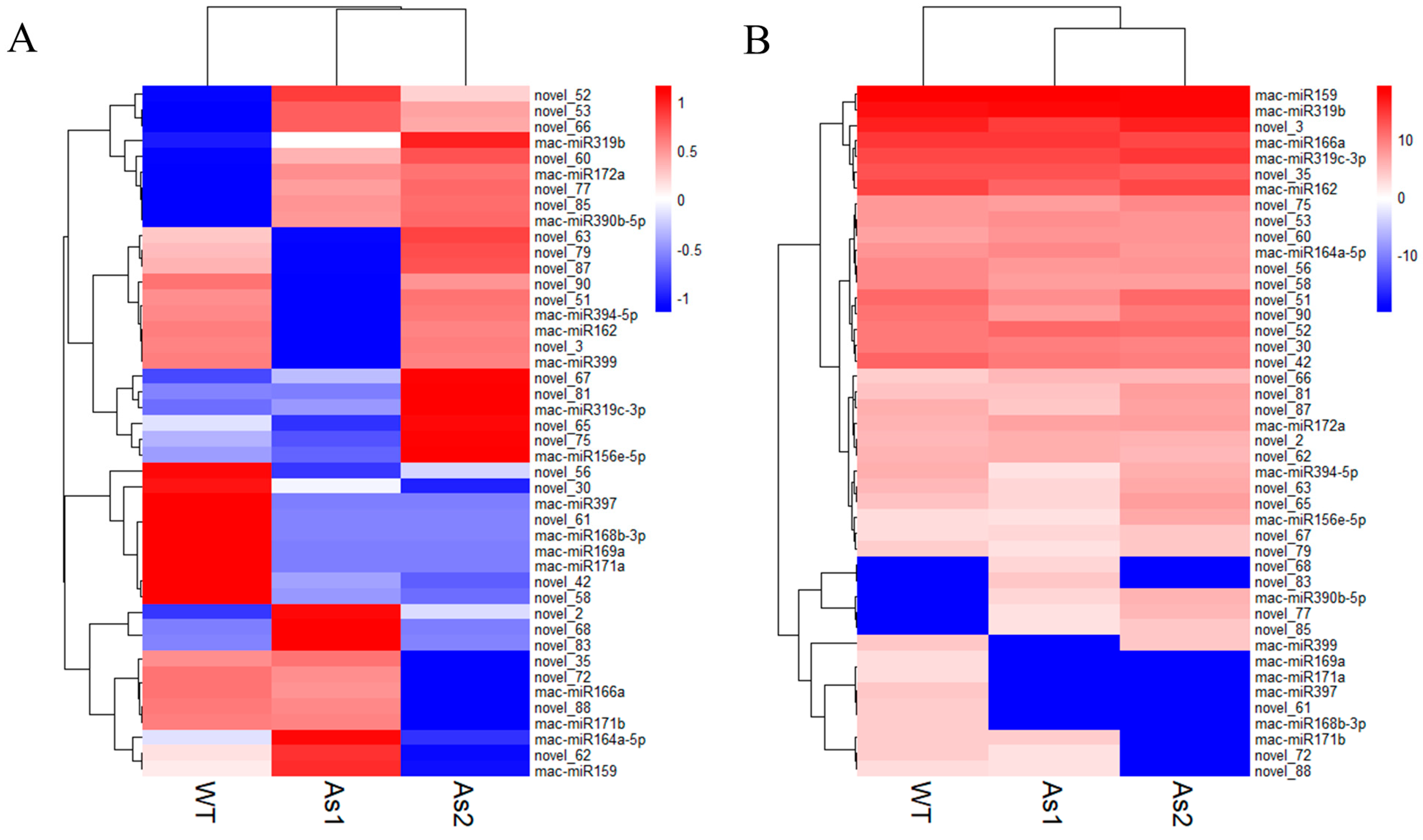
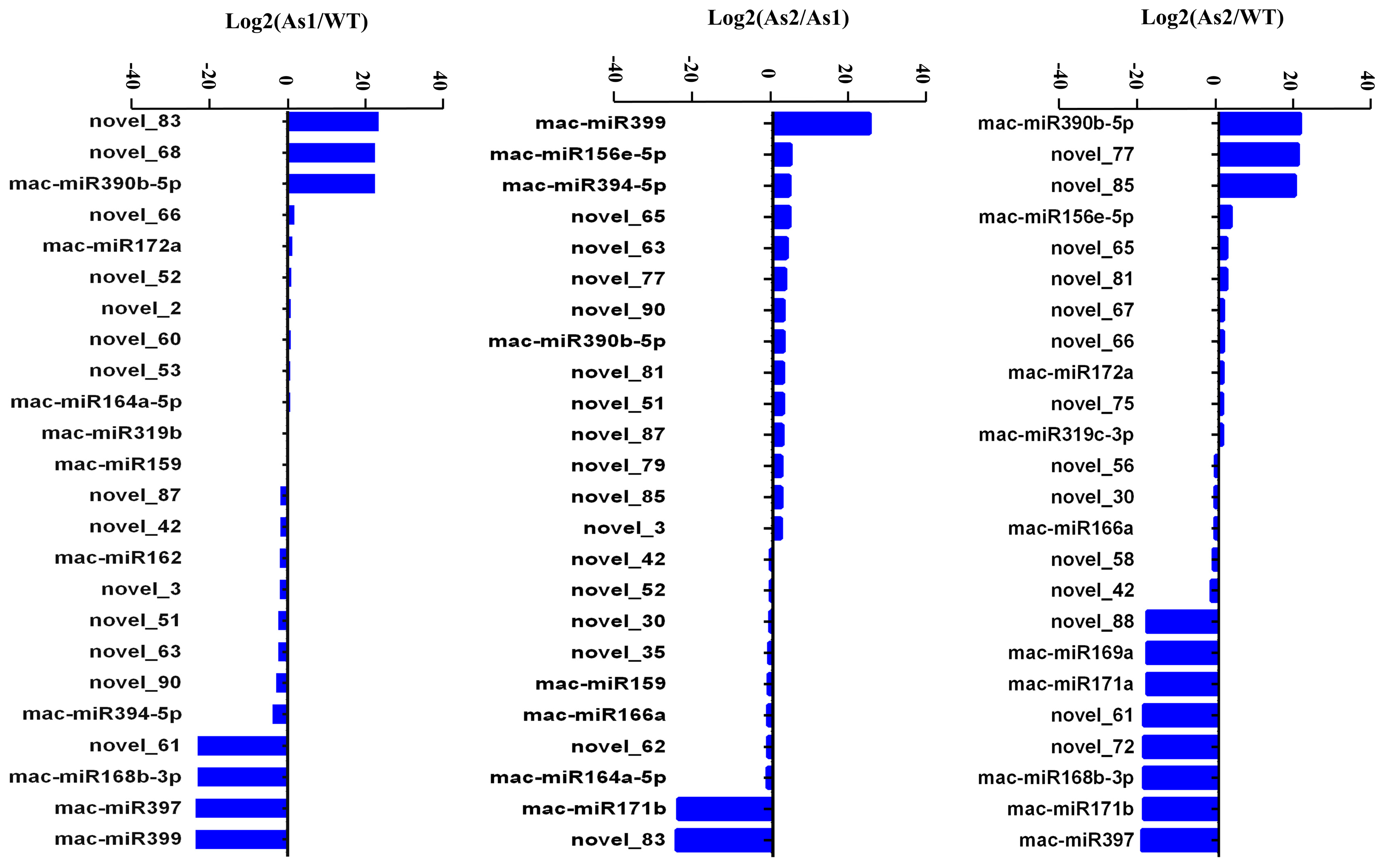
2.4. Differential Expression of MicroRNAs among Wild-Type and Two Transgenic Bananas
2.5. Expression Profiles of MicroRNA-Targeted Transcription Factor Genes in Mh-ACO1 and Mh-ACO2 RNAi Transgenic Bananas
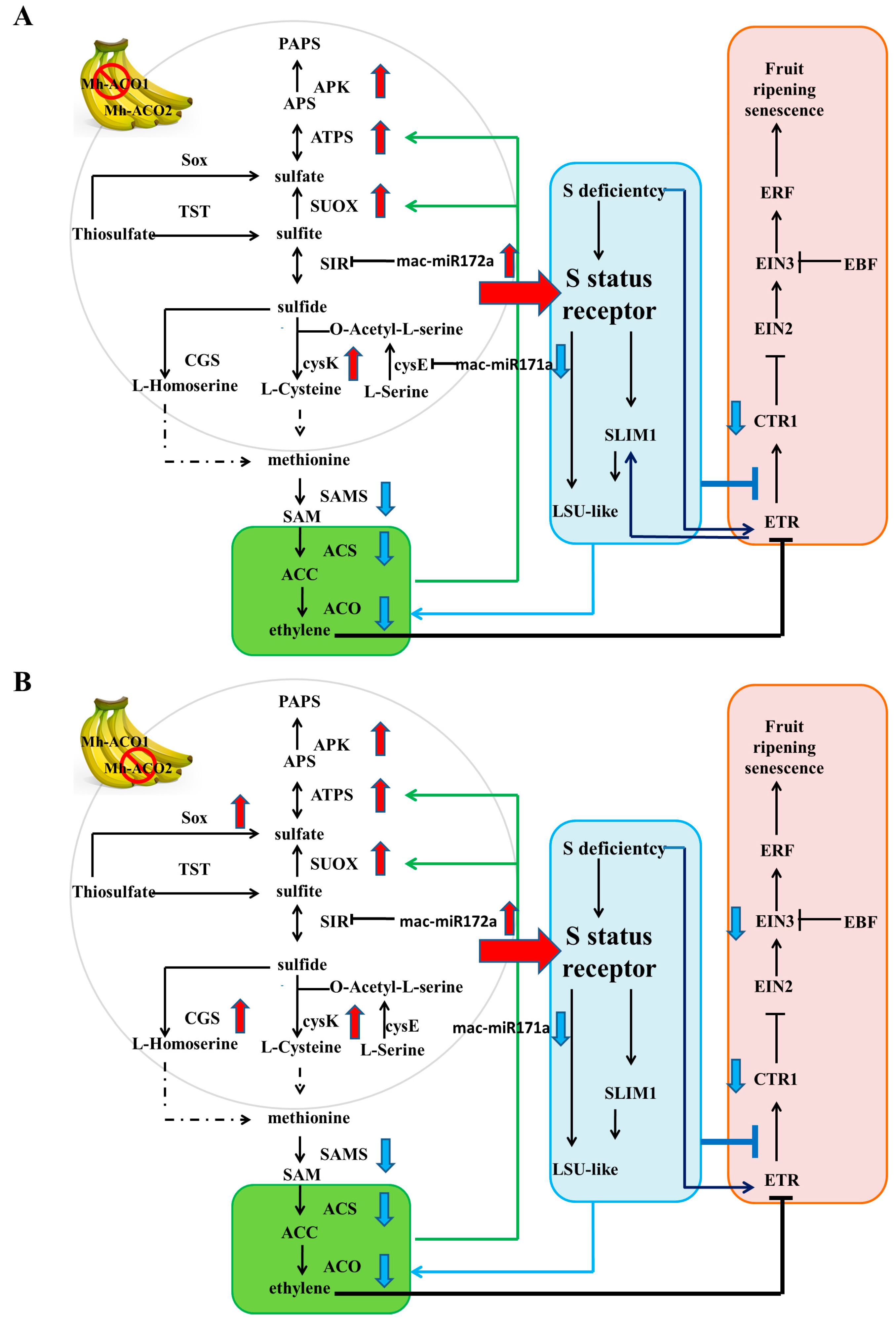
2.6. Feedback Regulation of Sulfur Metabolism Pathway, Ethylene Biosynthesis, and Signaling Pathway in Mh-ACO1 and Mh-ACO2 RNAi Transgenic Bananas
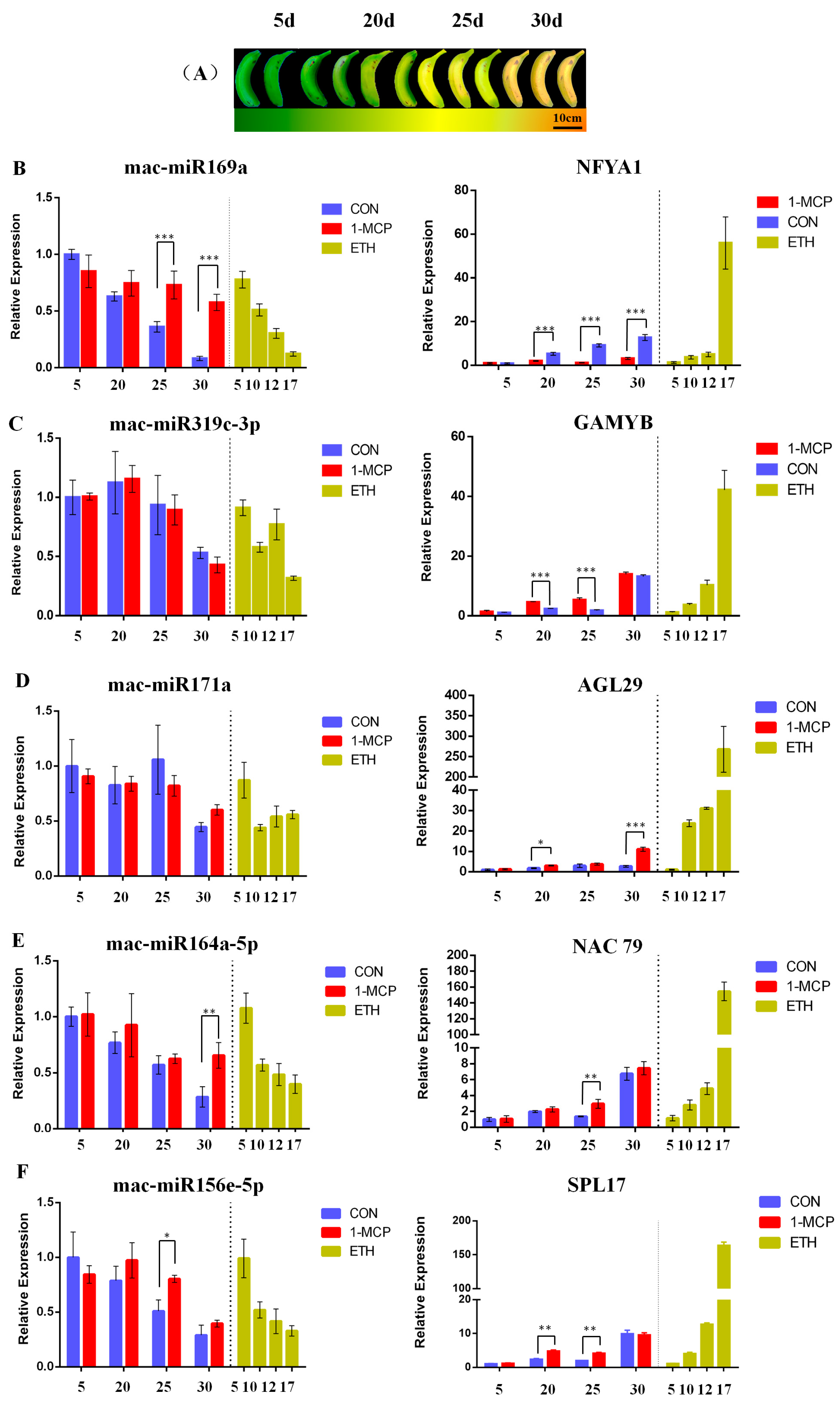
2.7. Differential Expression of MicroRNAs and Their Target Genes during Banana Fruit Ripening after Ethylene Treatments
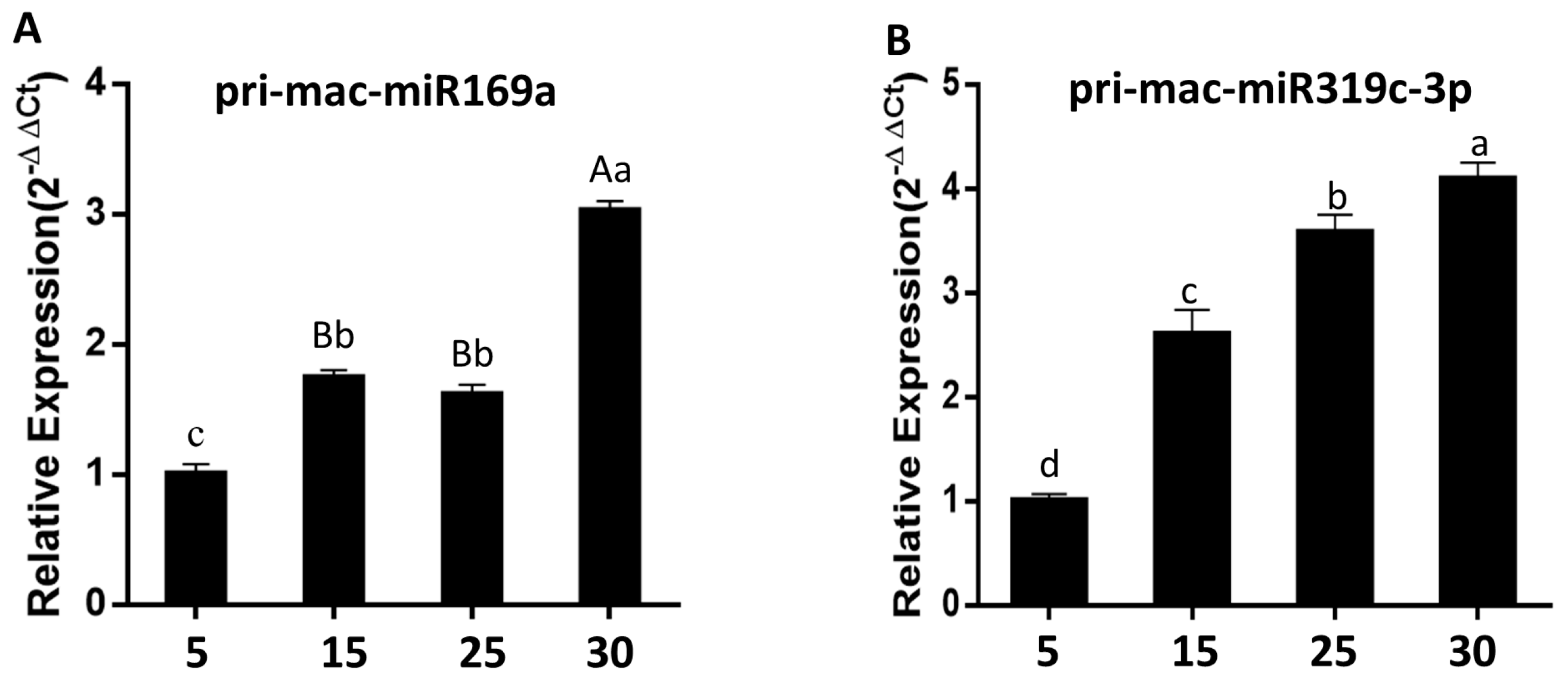
2.8. Cloning and Expression Analysis of Banana Pri-mac-miR169a and Pri-mac-miR319c-3p during Fruit Ripening
2.9. Intraspecific Cluster Analysis of Mature MicroRNAs Expression in Various Tissues of Bbanana
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation and Small RNA Sequencing
4.3. R Language Program for Heat Map Explanation
4.4. Chromosome Mapping and Phylogenetic Tree Construction of Banana ACC Oxidase Gene Family
4.5. 1-MCP and Ethylene Treatment of Banana Fruits
4.6. Analysis of the Expression Levels of MicroRNAs in Various Tissues of Bananas
4.7. Real-Time Fluorescence Quantitative PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Debat, H.J.; Ducasse, D.A. Plant microRNAs: Recent advances and future challenges. Plant Mol. Biol. Rep. 2014, 32, 1257–1269. [Google Scholar] [CrossRef]
- Chung, M.-Y.; Nath, U.K.; Vrebalov, J.; Gapper, N.; Lee, J.M.; Lee, D.-J.; Kim, C.K.; Giovannoni, J. Ectopic expression of miRNA172 in tomato (Solanum lycopersicum) reveals novel function in fruit development through regulation of an AP2 transcription factor. BMC Plant Biol. 2020, 20, 283. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ju, Z.; Cao, D.; Zhai, B.; Qin, G.; Zhu, H.; Fu, D.; Luo, Y.; Zhu, B. Micro RNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 2015, 13, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zou, W.; Xiao, Y.; Cheng, L.; Liu, Y.; Gao, S.; Shi, Z.; Jiang, Y.; Qi, M.; Xu, T.; et al. MicroRNA1917 targets CTR4 splice variants to regulate ethylene responses in tomato. J. Exp. Bot. 2018, 69, 1011–1025. [Google Scholar] [CrossRef]
- Chen, W.; Kong, J.; Lai, T.; Manning, K.; Wu, C.; Wang, Y.; Qin, C.; Li, B.; Yu, Z.; Zhang, X.; et al. Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci. Rep. 2015, 5, 7852. [Google Scholar] [CrossRef]
- Li, D.; Mou, W.; Luo, Z.; Li, L.; Limwachiranon, J.; Mao, L.; Ying, T. Developmental and stress regulation on expression of a novel miRNA, Fan-miR73, and its target ABI5 in strawberry. Sci. Rep. 2016, 6, 28385. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014, 55, 119–135. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Wu, Y.; Li, D.; Allan, A.C.; Yin, X. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020, 225, 1618–1634. [Google Scholar] [CrossRef]
- Wei, C.; Yang, R. Effect of over-expression miR319 on fruit quality of tomato. J. Anhui Agric. Sci. 2014, 42, 6150–6151. [Google Scholar] [CrossRef]
- Jia, X.; Shen, J.; Liu, H.; Li, F.; Ding, N.; Gao, C.; Pattanaik, S.; Patra, B.; Li, R.; Yuan, L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 2015, 242, 283–293. [Google Scholar] [CrossRef]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Zheng, H.; Dong, L.; Han, X.; Jin, H.; Yin, C.; Han, Y.; Li, B.; Qin, H.; Zhang, J.; Shen, Q.; et al. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol. 2020, 225, 2526–2541. [Google Scholar] [CrossRef]
- Chen, H.; Sun, J.; Li, S.; Cui, Q.; Zhang, H.; Xin, F.; Wang, H.; Lin, T.; Gao, D.; Wang, S.; et al. An ACC oxidase gene essential for cucumber carpel development. Mol. Plant 2016, 9, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.F.; Shan, W.; Liang, S.M.; Wu, C.J.; Wei, W.; Chen, J.Y.; Lu, W.J.; Kuang, J.F. MaBZR1/2 act as transcriptional repressors of ethylene biosynthetic genes in banana fruit. Physiol. Plant. 2019, 165, 555–568. [Google Scholar] [CrossRef]
- Wang, J.N.; Kuang, J.F.; Shan, W.; Chen, J.; Xie, H.; Lu, W.J.; Chen, J.W.; Chen, J.Y. Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep. 2012, 31, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Roy, S.; Nag, A.; Singh, S.K.; Sengupta, D.N. Characterization of an AGAMOUS-like MADS box protein, a probable constituent of flowering and fruit ripening regulatory system in banana. PLoS ONE 2012, 7, e44361. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Wang, J.; Zhang, J.; Miao, H.; Wang, Z.; Jia, C.; Zhang, J.; Xu, B.; Jin, Z. Transcription factor MaMADS36 plays a central role in regulating banana fruit ripening. J. Exp. Bot. 2021, 72, 7078–7091. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Qin, J.; Zheng, Q.; Ding, X.; Chen, W.; Lu, W.; Li, X.; Zhu, X. The involvement of the banana F-box protein MaEBF1 in regulating chilling-inhibited starch degradation through interaction with a MaNAC67-like protein. Biomolecules 2019, 9, 552. [Google Scholar] [CrossRef]
- Huang, F.C.; Do, Y.Y.; Huang, P.L. Genomic organization of a diverse ACC synthase gene family in banana and expression characteristics of the gene member involved in ripening of banana fruits. J. Agric. Food Chem. 2006, 54, 3859–3868. [Google Scholar] [CrossRef] [PubMed]
- Do, Y.Y.; Thay, T.S.; Chang, T.W.; Huang, P.L. Molecular cloning and characterization of a novel 1-aminocyclopropane-1-carboxylate oxidase gene involved in ripening of banana fruits. J. Agric. Food Chem. 2005, 53, 8239–8247. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.L.; Do, Y.Y.; Huang, F.C.; Thay, T.S.; Chang, T.W. Characterization and expression analysis of a banana gene encoding 1-aminocyclopropane-1-carboxylate oxidase. IUBMB Life 1997, 41, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Kuan, C.; Chiu, C.H.; Chen, X.J.; Do, Y.Y.; Huang, P.L. Global transcriptomic analysis of targeted silencing of two paralogous ACC oxidase genes in banana. Int. J. Mol. Sci. 2016, 17, 1632–1652. [Google Scholar] [CrossRef]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinf. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Tao, C.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Guo, A.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Osorio, S.; Bombarely, A.; Casañal, A.; Cruz-Rus, E.; Sánchez-Sevilla, J.F.; Amaya, I.; Giavalisco, P.; Fernie, A.R.; Botella, M.Á.; et al. Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening. New Phytol. 2015, 208, 482–496. [Google Scholar] [CrossRef]
- Wang, S.; Lu, G.; Hou, Z.; Luo, Z.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 2014, 65, 3005–3014. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.; Chen, L.; Xie, H.; Peng, H.; Xiao, Y.; Li, X.; Chen, W.; He, Q.; Chen, J.; et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, H.; Wall, M.M.; Yang, J. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 2020, 112, 2734–2747. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From data annotation to functional exploration of plant microRNAs. Nucleic Acids Res. 2022, 50, D1475–D1483. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B.C. Plant small RNAs: Their biogenesis, regulatory roles, and functions. Ann. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef] [PubMed]
- Qiang, Y.; Cui, X.; Shuai, L.; Gan, S.; Han, X.; Dou, D.; Hon-Ming, L. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef]
- Kawashima, C.G.; Berkowitz, O.; Hell, R.; Noji, M.; Saito, K. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol. 2005, 137, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Masood, A.; Khan, M.; Asgher, M.; Fatma, M.; Khan, N.A. Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal Behav 2013, 8, e22478. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA promoter binding protein–like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Araki, R.; Mermod, M.; Yamasaki, H.; Kamiya, T.; Fujiwara, T.; Shikanai, T. SPL7 locally regulates copper-homeostasis-related genes in Arabidopsis. J. Plant Physiol. 2018, 224, 137–143. [Google Scholar] [CrossRef]
- Ren, L.; Tang, G. Identification of sucrose-responsive microRNAs reveals sucrose-regulated copper accumulations in an SPL7-dependent and independent manner in Arabidopsis thaliana. Plant Sci. 2012, 187, 59–68. [Google Scholar] [CrossRef]
- Ba, L.J.; Wei, S.; Xiao, Y.Y.; Chen, J.Y.; Lu, W.-J.; Kuang, J.F. A ripening-induced transcription factor MaBSD1 interacts with promoters of MaEXP1/2 from banana fruit. Plant Cell Rep. 2014, 33, 1913–1920. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, S.; Feng, G.; Yi, H. Comparative analysis of miRNAs and their target transcripts between a spontaneous late-ripening sweet orange mutant and its wild-type using small RNA and degradome sequencing. Front. Plant Sci. 2016, 7, 1416. [Google Scholar] [CrossRef] [PubMed]
- Min, C.; Hui, C.; Hai, B.; Peng, H.; Yan-Wei, W. Advances in the research of miRNA promoters in plants. China Biotechnol. 2016, 36, 125–131. [Google Scholar] [CrossRef]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Megraw, M.; Baev, V.; Rusinov, V.; Jensen, S.T.; Kalantidis, K.; Hatzigeorgiou, A.G. MicroRNA promoter element discovery in Arabidopsis. RNA 2006, 12, 1612–1619. [Google Scholar] [CrossRef]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef]
- Hu, H.-L.; Do, Y.-Y.; Huang, P.-L. Expression profiles of a MhCTR1 gene in relation to banana fruit ripening. Plant Physiol. Biochem. 2012, 56, 47–55. [Google Scholar] [CrossRef]
- Commonwealth Scientific and Industrial Research Organization (CSIRO). Banana Ripening Guide; Division of Food Research, Circular; CSIRO: Canberra, Australia, 1972; Volume 8, pp. 1–12. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Barman, P.; Choudhary, A.K.; Geeta, R. A modified protocol yields high-quality RNA from highly mucilaginous Dioscorea tubers. 3 Biotech. 2017, 7, 150. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinf. 2012, 13, 140. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27. 3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene ontology testing for RNA-seq datasets. R Bioconductor 2012, 8, 1–25. [Google Scholar]
- Chao, J.; Kong, Y.; Wang, Q.; Sun, Y.; Liu, G. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 2015, 37, 91. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]




| Sample | WT | As1 | As2 |
|---|---|---|---|
| total_reads | 7,663,656 (100.00%) | 8,864,286 (100.00%) | 8,402,017 (100.00%) |
| N% > 10% | 899 (0.01%) | 296 (0.00%) | 1190 (0.01%) |
| low quality | 381 (0.00%) | 712 (0.01%) | 7365 (0.09%) |
| 5_adapter_contamine | 4189 (0.05%) | 2226 (0.03%) | 2212 (0.03%) |
| 3_adapter_null or insert_null | 737,544 (9.62%) | 513,274 (5.79%) | 924,855 (11.01%) |
| with ployA/T/G/C | 8022 (0.10%) | 10,013 (0.11%) | 9086 (0.11%) |
| clean reads | 6,912,621 (90.20%) | 8,337,765 (94.06%) | 7,457,309 (88.76%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Lai, Z.; Do, Y.-Y.; Huang, P.-L. Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas. Plants 2023, 12, 3414. https://doi.org/10.3390/plants12193414
Xia Y, Lai Z, Do Y-Y, Huang P-L. Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas. Plants. 2023; 12(19):3414. https://doi.org/10.3390/plants12193414
Chicago/Turabian StyleXia, Yan, Zhongxiong Lai, Yi-Yin Do, and Pung-Ling Huang. 2023. "Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas" Plants 12, no. 19: 3414. https://doi.org/10.3390/plants12193414
APA StyleXia, Y., Lai, Z., Do, Y.-Y., & Huang, P.-L. (2023). Characterization of MicroRNAs and Gene Expression in ACC Oxidase RNA Interference-Based Transgenic Bananas. Plants, 12(19), 3414. https://doi.org/10.3390/plants12193414









