Phyllosphere Microbiome in Plant Health and Disease
Abstract
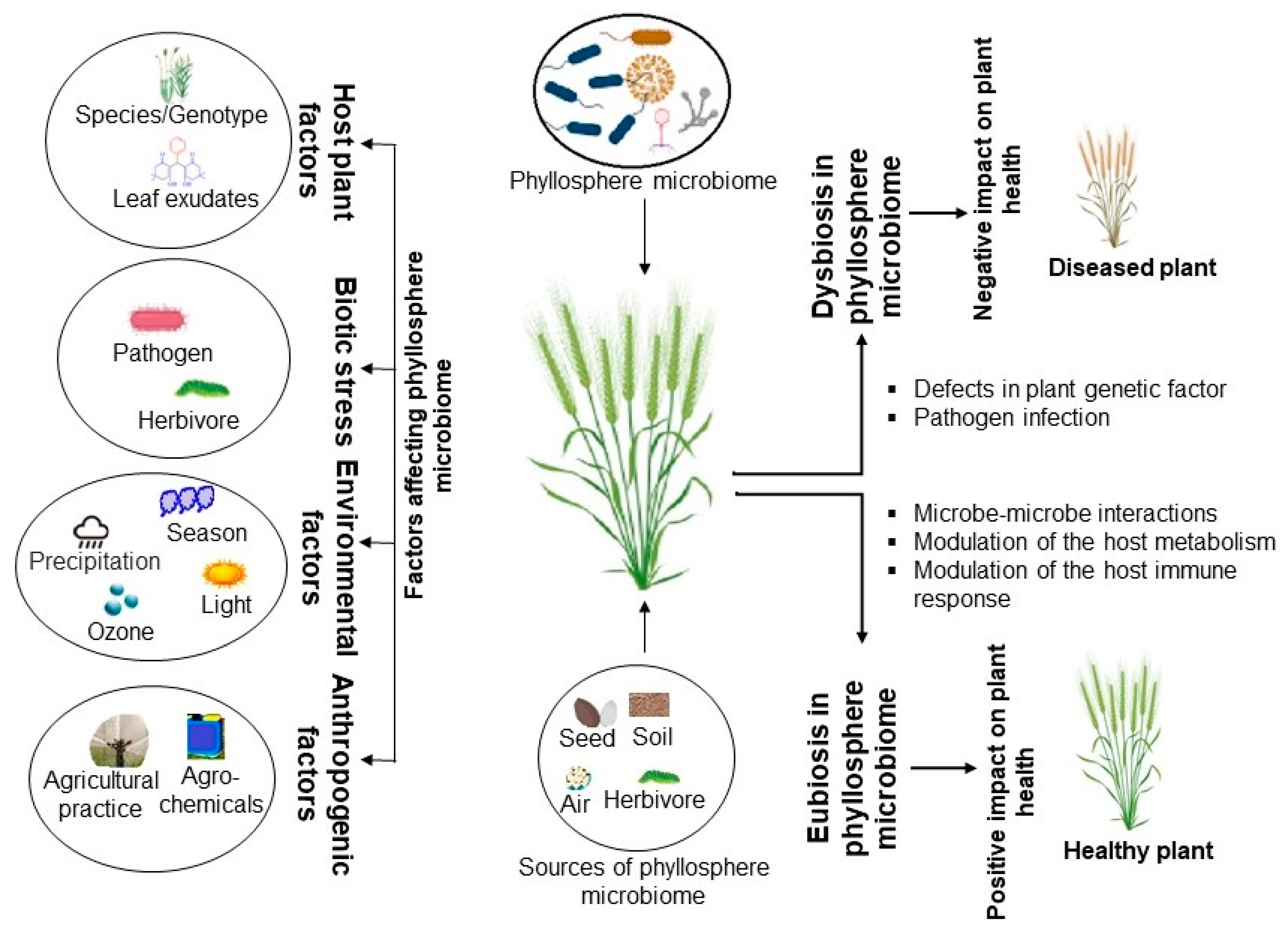
:1. Introduction
2. Mechanisms of Phyllosphere Microbiome Assembly
2.1. Host Genotype
2.2. Leaf Exudates
2.3. Environmental Factors
2.4. Anthropogenic Factors
2.5. Microbe–Microbe Interactions and Herbivores Impact
3. Role of the Phyllosphere Microbiome in Plant Defence
3.1. Microbe–Microbe Interactions
3.2. Modulation of the Host Metabolism
3.3. Modulation of the Host Immune Response
4. Future Research Topics
- (I)
- How does a plant regulate phyllosphere bacteriophage communities?
- (II)
- Does the “cry for help” strategy apply in the phyllosphere?
- (III)
- How does a plant maintain the phyllosphere microbial homeostasis?
- (IV)
- How does a disturbed phyllosphere microbiome affect the host plant?
- (V)
- What are the major methodological constraints for analyzing the phyllosphere microbiome?
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Leveau, J.H.J. Microbial communities in the phyllosphere. In Biology of the Plant Cuticle; Riederer, M., Müller, C., Eds.; Blackwell Publishing: Oxford, UK, 2006; pp. 334–367. [Google Scholar]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N.P. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Bright, M.; Bulgheresi, S. A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 2010, 8, 218–230. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of plant fraction, soil, and plant species on microbiota: A multikingdom comparison. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.-H.; Cheng, H.-P.; Jing, Y.-X.; Yanni, Y.G.; Dazzo, F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef]
- Vogel, C.; Bodenhausen, N.; Gruissem, W.; Vorholt, J.A. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016, 212, 192–207. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Singh, B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020, 25, 841–844. [Google Scholar] [CrossRef]
- Xu, N.; Zhao, Q.; Zhang, Z.; Zhang, Q.; Wang, Y.; Qin, G.; Ke, M.; Qiu, D.; Peijnenburg, W.J.G.M.; Lu, T.; et al. Phyllosphere Microorganisms: Sources, Drivers, and Their Interactions with Plant Hosts. J. Agric. Food Chem. 2022, 70, 4860–4870. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-L.; Hu, H.-W.; He, Z.-Y.; Cui, L.; Zhu, Y.-G.; He, J.-Z. Potential of indigenous crop microbiomes for sustainable agriculture. Nat. Food 2021, 2, 233–240. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef]
- Lambais, M.R.; Barrera, S.E.; Santos, E.C.; Crowley, D.E.; Jumpponen, A. Phyllosphere metaproteomes of trees from the Brazilian Atlantic forest show high levels of functional redundancy. Microb. Ecol. 2017, 73, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J. A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome. Curr. Opin. Microbiol. 2019, 49, 41–49. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Chen, T.; Wang, W.; Liu, G.; Zhang, W.; Li, S.; Wang, M.; Zhao, C.; Zhou, H. Plant phenotypic traits eventually shape its microbiota: A common garden test. Front. Microbiol. 2018, 9, 2479. [Google Scholar] [CrossRef]
- Reisberg, E.E.; Hildebrandt, U.; Riederer, M.; Hentschel, U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS ONE 2013, 8, e78613. [Google Scholar] [CrossRef]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef] [PubMed]
- Beilsmith, K.; Thoen, M.P.M.; Brachi, B.; Gloss, A.D.; Khan, M.H.; Bergelson, J. Genome-wide association studies on the phyllosphere microbiome: Embracing complexity in host–microbe interactions. Plant J. 2019, 97, 164–181. [Google Scholar] [CrossRef]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Metraux, J.-P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Liu, W.; Ke, M.; Qu, Q.; Zhou, Z.; Lu, T.; Qian, H. Phyllosphere bacterial assemblage is affected by plant genotypes and growth stages. Microbiol. Res. 2021, 248, 126743. [Google Scholar] [CrossRef] [PubMed]
- Ryffel, F.; Helfrich, E.J.N.; Kiefer, P.; Peyriga, L.; Portais, J.-C.; Piel, J.; Vorholt, J.A. Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 2016, 10, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; von Mering, C.; Vorholt, J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef]
- Kemen, A.C.; Agler, M.T.; Kemen, E. Host–microbe and microbe–microbe interactions in the evolution of obligate plant parasitism. New Phytol. 2015, 206, 1207–1228. [Google Scholar] [CrossRef]
- Xu, P.; Fan, X.; Mao, Y.; Cheng, H.; Xu, A.; Lai, W.; Lv, T.; Hu, Y.; Nie, Y.; Zheng, X. Temporal metabolite responsiveness of microbiota in the tea plant phyllosphere promotes continuous suppression of fungal pathogens. J. Adv. Res. 2022, 39, 49–60. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, B.; Pan, Y.; Tao, S.; Zhang, N. Metabolite-Mediated Responses of Phyllosphere Microbiota to Rust Infection in Two Malus Species. Microbiol. Spectr. 2023, 11, e03831-22. [Google Scholar] [CrossRef]
- Gupta, R.; Elkabetz, D.; Leibman-Markus, M.; Sayas, T.; Schneider, A.; Jami, E.; Kleiman, M.; Bar, M. Cytokinin drives assembly of the phyllosphere microbiome and promotes disease resistance through structural and chemical cues. ISME J. 2022, 16, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Graindorge, S.; Villette, C.; Koechler, S.; Groh, C.; Comtet-Marre, S.; Mercier, P.; Magerand, R.; Peyret, P.; Heintz, D.; Schaller, H.; et al. The Arabidopsis thaliana–Streptomyces Interaction Is Controlled by the Metabolic Status of the Holobiont. Int. J. Mol. Sci. 2022, 23, 12952. [Google Scholar]
- Xu, N.; Qu, Q.; Zhang, Z.; Yuan, W.; Cui, H.; Shen, Y.; Lin, W.; Lu, T.; Qian, H. Effects of residual S-metolachlor in soil on the phyllosphere microbial communities of wheat (Triticum aestivum L.). Sci. Total Environ. 2020, 748, 141342. [Google Scholar] [CrossRef]
- Aydogan, E.L.; Budich, O.; Hardt, M.; Choi, Y.H.; Jansen-Willems, A.B.; Moser, G.; Müller, C.; Kämpfer, P.; Glaeser, S.P. Global warming shifts the composition of the abundant bacterial phyllosphere microbiota as indicated by a cultivation-dependent and-independent study of the grassland phyllosphere of a long-term warming field experiment. FEMS Microbiol. Ecol. 2020, 96, fiaa087. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.D.; Lindow, S.E. Effect of plant species and environmental conditions on ice nucleation activity of Pseudomonas syringae on leaves. Appl. Environ. Microbiol. 1988, 54, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Zhu, C.; Alam, M.S.; Tokida, T.; Sakai, H.; Nakamura, H.; Usui, Y.; Zhu, J.; Hasegawa, T.; Jia, Z. Response of soil, leaf endosphere and phyllosphere bacterial communities to elevated CO2 and soil temperature in a rice paddy. Plant Soil 2015, 392, 27–44. [Google Scholar]
- Godon, J.J.; Galès, A.; Latrille, E.; Ouichanpagdee, P.; Seyer, J.-P. An “overlooked” habitat for thermophilic bacteria: The phyllosphere. BioDiscovery 2020, 23, e47033. [Google Scholar] [CrossRef]
- Chen, Q.L.; Hu, H.W.; Yan, Z.Z.; Li, C.Y.; Nguyen, B.A.T.; Zhu, Y.G.; He, J.Z. Precipitation increases the abundance of fungal plant pathogens in Eucalyptus phyllosphere. Environ. Microbiol. 2021, 23, 7688–7700. [Google Scholar] [CrossRef]
- Alsanius, B.W.; Bergstrand, K.-J.; Hartmann, R.; Gharaie, S.; Wohanka, W.; Dorais, M.; Rosberg, A.K. Ornamental flowers in new light: Artificial lighting shapes the microbial phyllosphere community structure of greenhouse grown sunflowers (Helianthus annuus L.). Sci. Hortic. 2017, 216, 234–247. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Finkel, O.M.; Burch, A.Y.; Lindow, S.E.; Post, A.F.; Belkin, S. Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Environ. Microbiol. 2011, 77, 7647–7655. [Google Scholar] [CrossRef]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef]
- Li, P.; Wu, X.; Gao, F. Ozone pollution, water deficit stress and time drive poplar phyllospheric bacterial community structure. Ecotoxicol. Environ. Saf. 2023, 262, 115148. [Google Scholar] [CrossRef]
- Qiu, D.; Ye, Y.; Ke, M.; Xu, N.; Zhang, Z.; Zhang, F.; Kang, J.; Yu, Y.; Lu, T.; Qian, H. Effects of chiral herbicide dichlorprop on Arabidopsis thaliana metabolic profile and its implications for microbial communities in the phyllosphere. Environ. Sci. Pollut. Res. 2022, 29, 28256–28266. [Google Scholar] [CrossRef]
- Xiang, Q.; Chen, Q.-L.; Zhu, D.; Yang, X.-R.; Qiao, M.; Hu, H.-W.; Zhu, Y.-G. Microbial functional traits in phyllosphere are more sensitive to anthropogenic disturbance than in soil. Environ. Pollut. 2020, 265, 114954. [Google Scholar] [CrossRef]
- Xiong, C.; He, J.Z.; Singh, B.K.; Zhu, Y.G.; Wang, J.T.; Li, P.P.; Zhang, Q.B.; Han, L.L.; Shen, J.P.; Ge, A.H. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 2021, 23, 1907–1924. [Google Scholar] [CrossRef]
- Berg, M.; Koskella, B. Nutrient- and Dose-Dependent Microbiome-Mediated Protection against a Plant Pathogen. Curr. Biol. 2018, 28, 2487–2492.e2483. [Google Scholar] [CrossRef]
- Wu, P.-H.; Chang, H.-X.; Shen, Y.-M. Effects of synthetic and environmentally friendly fungicides on powdery mildew management and the phyllosphere microbiome of cucumber. PLoS ONE 2023, 18, e0282809. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Messier, C.; Kembel, S. Tree Leaf Bacterial Community Structure and Diversity Differ along a Gradient of Urban Intensity. mSystems 2017, 2, e00087-17. [Google Scholar] [CrossRef]
- Espenshade, J.; Thijs, S.; Gawronski, S.; Bové, H.; Weyens, N.; Vangronsveld, J. Influence of Urbanization on Epiphytic Bacterial Communities of the Platanus × hispanica Tree Leaves in a Biennial Study. Front. Microbiol. 2019, 10, 675. [Google Scholar] [CrossRef]
- Smets, W.; Wuyts, K.; Oerlemans, E.; Wuyts, S.; Denys, S.; Samson, R.; Lebeer, S. Impact of urban land use on the bacterial phyllosphere of ivy (Hedera sp.). Atmos. Environ. 2016, 147, 376–383. [Google Scholar] [CrossRef]
- Hentschel, U.; Steinert, M.; Hacker, J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000, 8, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.T.; Whiteman, N.K. Insect herbivory reshapes a native leaf microbiome. Nat. Ecol. Evol. 2020, 4, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Müller, M.; Behrendt, U.; Stadler, B. Diversity of culturable phyllosphere bacteria on beech and oak: The effects of lepidopterous larvae. Microbiol. Res. 2003, 158, 291–297. [Google Scholar] [CrossRef]
- Smets, W.; Koskella, B. Microbiome: Insect herbivory drives plant phyllosphere dysbiosis. Curr. Biol. 2020, 30, R412–R414. [Google Scholar] [CrossRef]
- Tucker, C.M.; Fukami, T. Environmental variability counteracts priority effects to facilitate species coexistence: Evidence from nectar microbes. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132637. [Google Scholar] [CrossRef]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef]
- Morella, N.; Weng, F.; Joubert, P.; Metcalf, C.J.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2019, 117, 201908600. [Google Scholar] [CrossRef]
- Harsonowati, W.; Astuti, R.I.; Wahyudi, A.T. Leaf blast disease reduction by rice-phyllosphere actinomycetes producing bioactive compounds. J. Gen. Plant Pathol. 2017, 83, 98–108. [Google Scholar] [CrossRef]
- Fan, X.; Matsumoto, H.; Wang, Y.; Hu, Y.; Liu, Y.; Fang, H.; Nitkiewicz, B.; Lau, S.Y.L.; Wang, Q.; Fang, H.; et al. Microenvironmental Interplay Predominated by Beneficial Aspergillus Abates Fungal Pathogen Incidence in Paddy Environment. Environ. Sci. Technol. 2019, 53, 13042–13052. [Google Scholar] [CrossRef]
- Liu, X.; Matsumoto, H.; Lv, T.; Zhan, C.; Fang, H.; Pan, Q.; Xu, H.; Fan, X.; Chu, T.; Chen, S. Phyllosphere microbiome induces host metabolic defence against rice false-smut disease. Nat. Microbiol. 2023, 8, 1419–1433. [Google Scholar] [CrossRef] [PubMed]
- Sartori, M.; Nesci, A.; García, J.; Passone, M.A.; Montemarani, A.; Etcheverry, M. Efficacy of epiphytic bacteria to prevent northern leaf blight caused by Exserohilum turcicum in maize. Rev. Argent. Microbiol. 2017, 49, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Su, B.; Chen, X.; Xie, S.; Gu, S.; Wang, Q.; Huang, D.; Jiang, H. An endophytic bacterial strain isolated from Eucommia ulmoides inhibits southern corn leaf blight. Front. Microbiol. 2017, 8, 903. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Schenk, P.; Dart, P. Phyllosphere bacterial strains Rhizobium b1 and Bacillus subtilis b2 control tomato leaf diseases caused by Pseudomonas syringae pv. tomato and Alternaria solani. J. Appl. Microbiol. 2023, 134, lxad139. [Google Scholar] [CrossRef]
- Kefi, A.; Slimene, I.B.; Karkouch, I.; Rihouey, C.; Azaeiz, S.; Bejaoui, M.; Belaid, R.; Cosette, P.; Jouenne, T.; Limam, F. Characterization of endophytic Bacillus strains from tomato plants (Lycopersicon esculentum) displaying antifungal activity against Botrytis cinerea Pers. World J. Microbiol. Biotechnol. 2015, 31, 1967–1976. [Google Scholar] [CrossRef]
- Chaouachi, M.; Marzouk, T.; Jallouli, S.; Elkahoui, S.; Gentzbittel, L.; Ben, C.; Djébali, N. Activity assessment of tomato endophytic bacteria bioactive compounds for the postharvest biocontrol of Botrytis cinerea. Postharvest Biol. Technol. 2021, 172, 111389. [Google Scholar] [CrossRef]
- Qin, C.; Tao, J.; Liu, T.; Liu, Y.; Xiao, N.; Li, T.; Gu, Y.; Yin, H.; Meng, D. Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express 2019, 9, 42. [Google Scholar] [CrossRef]
- Michavila, G.; Adler, C.; De Gregorio, P.R.; Lami, M.J.; Caram Di Santo, M.C.; Zenoff, A.M.; de Cristobal, R.E.; Vincent, P.A. Pseudomonas protegens CS 1 from the lemon phyllosphere as a candidate for citrus canker biocontrol agent. Plant Biol. 2017, 19, 608–617. [Google Scholar] [CrossRef]
- Eitzen, K.; Sengupta, P.; Kroll, S.; Kemen, E.; Doehlemann, G. A fungal member of the Arabidopsis thaliana phyllosphere antagonizes Albugo laibachii via a GH25 lysozyme. eLife 2021, 10, e65306. [Google Scholar] [CrossRef]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Someya, N.; Ikeda, T. Novel N-acylhomoserine lactone-degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 2009, 73, 2124–2127. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-J.; Chou, H.-P.; Huang, J.-W.; Deng, W.-L. Genomic and biochemical characterization of antifungal compounds produced by Bacillus subtilis PMB102 against Alternaria brassicicola. Microbiol. Res. 2021, 251, 126815. [Google Scholar] [CrossRef] [PubMed]
- Shade, A. Diversity is the question, not the answer. ISME J. 2017, 11, 1–6. [Google Scholar] [CrossRef]
- Yang, H.; Li, J.; Xiao, Y.; Gu, Y.; Liu, H.; Liang, Y.; Liu, X.; Hu, J.; Meng, D.; Yin, H. An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 2017, 8, 2179. [Google Scholar] [CrossRef]
- D’Alessandro, M.; Erb, M.; Ton, J.; Brandenburg, A.; Karlen, D.; Zopfi, J.; Turlings, T.C.J. Volatiles produced by soil-borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant Cell Environ. 2014, 37, 813–826. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Vogel, C.M.; Ueoka, R.; Schäfer, M.; Ryffel, F.; Müller, D.B.; Probst, S.; Kreuzer, M.; Piel, J.; Vorholt, J.A. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat. Microbiol. 2018, 3, 909–919. [Google Scholar] [CrossRef]
- Ma, A.; Lv, D.; Zhuang, X.; Zhuang, G. Quorum Quenching in Culturable Phyllosphere Bacteria from Tobacco. Int. J. Mol. Sci. 2013, 14, 14607–14619. [Google Scholar] [CrossRef]
- Zhang, W.; Peng, Q.; Chen, L.; Gu, Z.; Liu, Z.; Zhang, D.; Cheng, J.E.; Zheng, L.; Chen, A.; Liu, Y. Quorum Sensing Is Required for the Colony Establishment of a Plant Phyllosphere Bacterium Rhodopseudomonas palustris Strain GJ-22. Appl. Environ. Microbiol. 2023, 89, e0048723. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.r.; Guenther, A.; Llusia, J.; Rico, L.; Terradas, J.; Farré-Armengol, G.; Filella, I.; Parella, T.; et al. Shifts in plant foliar and floral metabolomes in response to the suppression of the associated microbiota. BMC Plant Biol. 2016, 16, 78. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Tan, X.; Li, C.; Zhang, D.; Cheng, J.; Zhang, S.; Zhou, X.; Yan, Q.; Peng, J.; Zhang, Z.; et al. Photosynthetic bacterium Rhodopseudomonas palustris GJ-22 induces systemic resistance against viruses. Microb. Biotechnol. 2017, 10, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Bozsóki, Z.; Gysel, K.; Hansen, S.; Lironi, D.; Krönauer, C.; Feng, F.; Jong, N.; Vinther, M.; Kamble, M.; Thygesen, M.; et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 2020, 369, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.; Kiefer, P.; Field, C.; Hemmerle, L.; Bortfeld-Miller, M.; Emmenegger, B.; Schäfer, M.; Pfeilmeier, S.; Sunagawa, S.; Vogel, C.; et al. A general non-self response as part of plant immunity. Nat. Plants 2021, 7, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, B.; Biosca, E.G. Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Bhunchoth, A.; Phironrit, N.; Leksomboon, C.; Chatchawankanphanich, O.; Kotera, S.; Narulita, E.; Kawasaki, T.; Fujie, M.; Yamada, T. Isolation of R alstonia solanacearum-infecting bacteriophages from tomato fields in C hiang M ai, T hailand, and their experimental use as biocontrol agents. J. Appl. Microbiol. 2015, 118, 1023–1033. [Google Scholar] [CrossRef]
- Morella, N.M.; Gomez, A.L.; Wang, G.; Leung, M.S.; Koskella, B. The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol. Ecol. 2018, 27, 2025–2038. [Google Scholar] [CrossRef]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef]
- Wen, T.; Zhao, M.; Yuan, J.; Kowalchuk, G.A.; Shen, Q. Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 2021, 3, 42–51. [Google Scholar] [CrossRef]
- Yuan, J.; Zhao, J.; Wen, T.; Zhao, M.; Li, R.; Goossens, P.; Huang, Q.; Bai, Y.; Vivanco, J.M.; Kowalchuk, G.A.; et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 2018, 6, 156. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Wu, G.; Feng, H.; Zhang, G.; Shen, Q.; Zhang, R. Identification of root-secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 2017, 30, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.M.; Potthoff, D.B.; Schäfer, M.; Barandun, N.; Vorholt, J.A. Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nat. Microbiol. 2021, 6, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Ehau-Taumaunu, H.; Hockett, K.L. Passaging phyllosphere microbial communities develop suppression towards bacterial speck disease in tomato. Phytobiomes J. 2022, 7, PBIOMES-05. [Google Scholar] [CrossRef]
- Li, P.-D.; Zhu, Z.-R.; Zhang, Y.; Xu, J.; Wang, H.; Wang, Z.; Li, H. The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome 2022, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Nomura, K.; Wang, X.; Sohrabi, R.; Xu, J.; Yao, L.; Paasch, B.C.; Ma, L.; Kremer, J.; Cheng, Y.; et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 2020, 580, 653–657. [Google Scholar] [CrossRef]
- Pfeilmeier, S.; Petti, G.C.; Bortfeld-Miller, M.; Daniel, B.; Field, C.M.; Sunagawa, S.; Vorholt, J.A. The plant NADPH oxidase RBOHD is required for microbiota homeostasis in leaves. Nat. Microbiol. 2021, 6, 852–864. [Google Scholar] [CrossRef]
- Das, B.; Nair, G.B. Homeostasis and dysbiosis of the gut microbiome in health and disease. J. Biosci. 2019, 44, 117. [Google Scholar] [CrossRef]
- Pfeilmeier, S.; Werz, A.; Ote, M.; Bortfeld-Miller, M.; Kirner, P.; Keppler, A.; Hemmerle, L.; Gaebelein, C.G.; Pestalozzi, C.M.; Vorholt, J.A. Dysbiosis of a leaf microbiome is caused by enzyme secretion of opportunistic Xanthomonas strains. bioRxiv 2023. [Google Scholar] [CrossRef]
- Seybold, H.; Demetrowitsch, T.J.; Hassani, M.A.; Szymczak, S.; Reim, E.; Haueisen, J.; Lübbers, L.; Rühlemann, M.; Franke, A.; Schwarz, K.; et al. A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nat. Commun. 2020, 11, 1910. [Google Scholar] [CrossRef]
- Suda, W.; Nagasaki, A.; Shishido, M. Powdery mildew-infection changes bacterial community composition in the phyllosphere. Microbes Environ. 2009, 24, 217–223. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.-C.; Cai, L.-T.; Li, W.; Pan, D.; Xiang, L.; Su, X.; Li, Z.; Adil, M.F.; Shamsi, I.H. Phyllospheric microbial composition and diversity of the tobacco leaves infected by Didymella segeticola. Front. Microbiol. 2021, 12, 699699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Luo, L.; Tan, X.; Kong, X.; Yang, J.; Wang, D.; Zhang, D.; Jin, D.; Liu, Y. Pumpkin powdery mildew disease severity influences the fungal diversity of the phyllosphere. PeerJ 2018, 6, e4559. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, Z.; Wang, P.; Han, Y.; Jin, D.; Su, P.; Tan, X.; Zhang, D.; Muhammad-Rizwan, H.; Lu, X.; et al. Variations in phyllosphere microbial community along with the development of angular leaf-spot of cucumber. AMB Express 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Nobori, T.; Wang, Y.; Wu, J.; Stolze, S.C.; Tsuda, Y.; Finkemeier, I.; Nakagami, H.; Tsuda, K. Multidimensional gene regulatory landscape of a bacterial pathogen in plants. Nat. Plants 2020, 6, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, L.; Zhang, M.; Liu, D.; Sun, H.; Wu, J.; Huo, Y.; Chen, X.; Fang, R. Quantitative and qualitative characterization of plant endo-bacteriome by plant DNA-free sequencing method. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Wicaksono, W.A.; Bergna, A.; Cernava, T.; Bergau, N.; Tissier, A.; Hause, B.; Berg, G. Trichomes form genotype-specific microbial hotspots in the phyllosphere of tomato. Environ. Microbiome 2020, 15, 17. [Google Scholar] [CrossRef]
- Bell, J.K.; Helgason, B.; Siciliano, S.D. Brassica napus phyllosphere bacterial composition changes with growth stage. Plant Soil 2021, 464, 501–516. [Google Scholar] [CrossRef]
- Taerum, S.J.; Steven, B.; Gage, D.; Triplett, L.R. Dominance of Ciliophora and Chlorophyta Among Phyllosphere Protists of Solanaceous Plants. Phytobiomes J. 2022, 7, PBIOMES-04. [Google Scholar] [CrossRef]
- Song, L.; Xie, K. Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing. Microbiome 2020, 8, 80. [Google Scholar] [CrossRef]
| Plant | Pathogen | Phyllo Microbe | Mechanisms | Reference |
|---|---|---|---|---|
| Oryza sativa | Pyricularia oryzae | Actinomycetes | Produce bioactive compounds | [60] |
| Magnaporthe oryzae | Aspergillus cvjetkovicii | Produces 2(3H)-benzofuranone and azuline, which suppress mycelial growth and appressorium formation | [61] | |
| Ustilaginoidea virens | Panicle microbes | Modulates the levels of branched-chain amino acids | [62] | |
| Zea mays | Exserohilum turcicum | Enterococcus, Corynebacterium, Pantoea and Bacillus | Unknown mechanism | [63] |
| Bacillus subtilis strain DZSY21 | Bipolaris maydis | Reduce infection, possibly using antifungal lipopeptides and induced systemic response | [64] | |
| Triticum aestivum | Fusarium gramineareum | Pseudomonas piscium | Compound secreted by the bacteria (phenazine-1-carboxamide) deregulates histone acetylation and suppress growth, virulence, and mycotoxin biosynthesis. | [65] |
| Solanum lycopersicum | Pseudomonas syringae pv. tomato and Alternaria solani | Rhizobium sp. and Bacillus subtilis | Produce protease and cellulase and induce salicylic acid (SA) immune response pathway | [66] |
| Botrytis cinerea | Bacillus sp. | Produce lipopeptides antibiotics belonging to fengycin, surfactin, iturina and bacillomycin D | [67] | |
| Botrytis cinerea | Enterobacter cloacae TR1 | Produces antifungal volatile compound 3-methylbutan-1-ol | [68] | |
| Nicotiana tabacum | Pseudomonas syringae pv. tabaci | Stenotrophomonas, Achromobacter, Enterobacter, Ochrobactrum, Pseudomonas, Bacillus, Alcaligenes, Pseudochrobactrum and Achromobacte | Increases the complexity of microbial networks in the phyllosphere and establishes a “spatial repellent barrier” against invading pathogens | [69] |
| Citrus limon | Xanthomonas citri ssp. Citri | Pseudomonas protegens CS1 | Inhibit pathogen by producing siderophore pyochelin | [70] |
| Arabidopsis thaliana | Albugo laibachii | Moesziomyces bullatus ex Albugo | GH25 hydrolase secreted by the commensal play a major role in pathogen defence | [71] |
| Sphingomonas melonis Fr1 | Pseudomonas syringae DC3000 | Activates defence genes to promote immunity against pathogen | [10] | |
| Pseudomonas syringae pv. tomato DC3000 | Sphingomonas | Substrate competition plays a role in plant protection | [72] | |
| Solanum tuberosum | Microbacterium testaceum | Pectobacterium carotovorum | Interfere with the N-acyl-homoserine lactone (AHL)-based quorum-sensing of the pathgoen | [73] |
| Brassica rapa | Alternaria brassicicola ABA-31 | Bacillus subtilis PMB102 | Production of antifungal metabolites | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Mandal, S.; Jeon, J. Phyllosphere Microbiome in Plant Health and Disease. Plants 2023, 12, 3481. https://doi.org/10.3390/plants12193481
De Mandal S, Jeon J. Phyllosphere Microbiome in Plant Health and Disease. Plants. 2023; 12(19):3481. https://doi.org/10.3390/plants12193481
Chicago/Turabian StyleDe Mandal, Surajit, and Junhyun Jeon. 2023. "Phyllosphere Microbiome in Plant Health and Disease" Plants 12, no. 19: 3481. https://doi.org/10.3390/plants12193481
APA StyleDe Mandal, S., & Jeon, J. (2023). Phyllosphere Microbiome in Plant Health and Disease. Plants, 12(19), 3481. https://doi.org/10.3390/plants12193481






