Development and Evaluation of an AxiomTM 60K SNP Array for Almond (Prunus dulcis)
Abstract
1. Introduction
2. Results–Discussion
2.1. Array Design
2.1.1. SNPs Array Selection
2.1.2. Distribution of Array SNPs
2.2. Validation of the Axiom 60K Almond Array
2.2.1. Validation of the Array on the Diversity Panel
2.2.2. Validation of the Three Additional SNPs
2.2.3. Validation of the Array with a F2 Progeny
2.3. Evaluation of the Error Rates in Different Analyses
2.3.1. Comparison of “Texas” Genotyping with the “Texas” Reference Genome
2.3.2. Comparison of Array Data from Three Replicates of the “Ferrastar” Variety
2.3.3. Comparison between Expected and Observed Homozygote F2 Data
3. Materials and Methods
3.1. Plant Material Resequenced
3.2. SNP Calling and Selection of SNPs for Array Development
3.3. Plant Material for the 60K SNP Array Validation
3.4. Genotyping and SNP Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-Generation Sequencing Technologies and Their Implications for Crop Genetics and Breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-Assisted Breeding in Fruit Trees. Breed. Sci. 2016, 66, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Verde, I.; Bassil, N.; Scalabrin, S.; Gilmore, B.; Lawley, C.; Gasic, K.; Micheletti, D.; Rosyara, U.; Cattonaro, F.; Vendramin, E.; et al. Development and Evaluation of a 9K SNP Array for Peach by Internationally Coordinated SNP Detection and Validation in Breeding Germplasm. PLoS ONE 2012, 7, e35668. [Google Scholar] [CrossRef]
- Gasic, K.; Linge, C.D.S.; Bianco, L.; Troggio, M.; Rossini, L.; Bassi, D.; Aranzana, M.J.; Arus, P.; Verde, I.; Peace, C.; et al. Development and Evaluation of a 9K SNP Addition to the Peach Ipsc 9K SNP Array V1. Hortscience 2019, 54, S188. [Google Scholar]
- Vanderzande, S.; Zheng, P.; Cai, L.; Barac, G.; Gasic, K.; Main, D.; Iezzoni, A.; Peace, C. The Cherry 6+9K SNP Array: A Cost-Effective Improvement to the Cherry 6K SNP Array for Genetic Studies. Sci. Rep. 2020, 10, 7613. [Google Scholar] [CrossRef]
- Bianco, L.; Cestaro, A.; Linsmith, G.; Muranty, H.; Denance, C.; Theron, A.; Poncet, C.; Micheletti, D.; Kerschbamer, E.; Di Pierro, E.; et al. Development and Validation of the Axiom (R) Apple480K SNP Genotyping Array. PLANT J. 2016, 86, 62–74. [Google Scholar] [CrossRef]
- Montanari, S.; Bianco, L.; Allen, B.; Martinez-Garcia, P.; Bassil, N.; Postman, J.; Knabel, M.; Kitson, B.; Deng, C.; Chagne, D.; et al. Development of a Highly Efficient Axiom (TM) 70 K SNP Array for Pyrus and Evaluation for High-Density Mapping and Germplasm Characterization. BMC Genom. 2019, 20, 331. [Google Scholar] [CrossRef]
- Li, X.; Singh, J.; Qin, M.; Li, S.; Zhang, X.; Zhang, M.; Khan, A.; Zhang, S.; Wu, J. Development of an Integrated 200K SNP Genotyping Array and Application for Genetic Mapping, Genome Assembly Improvement and Genome Wide Association Studies in Pear (Pyrus). Plant Biotechnol. J. 2019, 17, 1582–1594. [Google Scholar] [CrossRef]
- Marrano, A.; Martinez-Garcia, P.J.; Bianco, L.; Sideli, G.M.; Di Pierro, E.A.; Leslie, C.A.; Stevens, K.A.; Crepeau, M.W.; Troggio, M.; Langley, C.H.; et al. A New Genomic Tool for Walnut (Juglans Regia L.): Development and Validation of the High-Density Axiom (TM) J. Regia 700K SNP Genotyping Array. Plant Biotechnol. J. 2019, 17, 1027–1036. [Google Scholar] [CrossRef]
- Di Guardo, M.; Farneti, B.; Khomenko, I.; Modica, G.; Mosca, A.; Distefano, G.; Bianco, L.; Troggio, M.; Sottile, F.; La Malfa, S.; et al. Genetic Characterization of an Almond Germplasm Collection and Volatilome Profiling of Raw and Roasted Kernels. Hortic. Res. 2021, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Fernandez i Marti, A.; Frias, L.; et al. Transposons Played a Major Role in the Diversification between the Closely Related Almond and Peach Genomes: Results from the Almond Genome Sequence. Plant J. 2020, 101, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pérez, R.; Pavan, S.; Mazzeo, R.; Moldovan, C.; Aiese Cigliano, R.; Del Cueto, J.; Ricciardi, F.; Lotti, C.; Ricciardi, L.; Dicenta, F.; et al. Mutation of a BHLH Transcription Factor Allowed Almond Domestication. Science 2019, 364, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- D’Amico-Willman, K.; Sideli, G.; Allen, B.; Anderson, E.; Gradziel, T.; Fresnedo-Ramirez, J. Identification of Putative Markers of Non-Infectious Bud Failure in Almond [Prunus Dulcis (Mill.) DA Webb] Through Genome Wide DNA Methylation Profiling and Gene Expression Analysis in an Almond × Peach Hybrid Population. Front. Plant Sci. 2022, 13, 804145. [Google Scholar] [CrossRef]
- Donoso, J.M.; Picanol, R.; Serra, O.; Howad, W.; Alegre, S.; Arus, P.; Eduardo, I. Exploring Almond Genetic Variability Useful for Peach Improvement: Mapping Major Genes and QTLs in Two Interspecific Almond x Peach Populations. Mol. Breed. 2016, 36, 16. [Google Scholar] [CrossRef]
- Duval, H.; Van Ghelder, C.; Callot, C.; Esmenjaud, D. Characterization of the RMja Resistance Gene to Root-Knot Nematodes from the ‘Alnem’ Almond Rootstock. Acta Hortic. 2018, 1219, 325–330. [Google Scholar] [CrossRef]
- Kochba, J.; Spiegelroy, P. Alnem-1, Alnem-88, Alnem-201 Almonds—Nematode-Resistant Rootstock Seed Source. Hortscience 1976, 11, 270. [Google Scholar] [CrossRef]
- Saucet, S.B.; Van Ghelder, C.; Abad, P.; Duval, H.; Esmenjaud, D. Resistance to Root-Knot Nematodes Meloidogyne Spp. in Woody Plants. New Phytol. 2016, 211, 41–56. [Google Scholar] [CrossRef]
- Van Ghelder, C.; Esmenjaud, D.; Callot, C.; Dubois, E.; Mazier, M.; Duval, H. Ma Orthologous Genes in Prunus Spp. Shed Light on a Noteworthy NBS-LRR Cluster Conferring Differential Resistance to Root-Knot Nematodes. Front. Plant Sci. 2018, 9, 1269. [Google Scholar] [CrossRef]
- Pavan, S.; Delvento, C.; Mazzeo, R.; Ricciardi, F.; Losciale, P.; Gaeta, L.; D’Agostino, N.; Taranto, F.; Sánchez-Pérez, R.; Ricciardi, L.; et al. Almond Diversity and Homozygosity Define Structure, Kinship, Inbreeding, and Linkage Disequilibrium in Cultivated Germplasm, and Reveal Genomic Associations with Nut and Seed Weight. Hortic. Res. 2021, 8, 15. [Google Scholar] [CrossRef]
- Sanchez-Perez, R.; Howad, W.; Garcia-Mas, J.; Arus, P.; Martinez-Gomez, P.; Dicenta, F. Molecular Markers for Kernel Bitterness in Almond. Tree Genet. Genomes 2010, 6, 237–245. [Google Scholar] [CrossRef]
- Yang, S.; Fresnedo-Ramírez, J.; Sun, Q.; Manns, D.C.; Sacks, G.L.; Mansfield, A.K.; Luby, J.J.; Londo, J.P.; Reisch, B.I.; Cadle-Davidson, L.E. Next Generation Mapping of Enological Traits in an F2 Interspecific Grapevine Hybrid Family. PLoS ONE 2016, 11, e0149560. [Google Scholar] [CrossRef] [PubMed]
- Ooijen, J.; Ooijen, J.; van ‘t Verlaat, J.W.; Ooijen, J.; Tol, J.; Dale, J.; Buren, J.; Meer, J.M.; Krieken, J.V.; Ooijen, J.; et al. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. 2006. Available online: https://www.kyazma.nl/index.php/JoinMap/ (accessed on 13 July 2013).
- Donoso, J.M.; Eduardo, I.; Picanol, R.; Batlle, I.; Howad, W.; Aranzana, M.J.; Arus, P. High-Density Mapping Suggests Cytoplasmic Male Sterility with Two Restorer Genes in Almond * Peach Progenies. Hortic. Res. 2015, 2, 15016. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, S.N.; March, T.J.; Wirthensohn, M.G.; Arús, P.; Walker, A.R.; Mather, D.E. Genotyping by Sequencing in Almond: SNP Discovery, Linkage Mapping, and Marker Design. G3 Genes Genomes Genet. 2018, 8, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Guajardo, V.; Martínez-García, P.J.; Solís, S.; Calleja-Satrustegui, A.; Saski, C.; Moreno, M.Á. QTLs Identification for Iron Chlorosis in a Segregating Peach–Almond Progeny through Double-Digest Sequence-Based Genotyping (SBG). Front. Plant Sci. 2022, 13, 872208. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fu, J.; Xu, Y.; Zhang, J.; Ren, F.; Zhao, H.; Tian, S.; Guo, W.; Tu, X.; Zhao, J. Genome Re-Sequencing Reveals the Evolutionary History of Peach Fruit Edibility. Nat. Commun. 2018, 9, 5404. [Google Scholar] [CrossRef]
- Koepke, T.; Schaeffer, S.; Harper, A.; Dicenta, F.; Edwards, M.; Henry, R.J.; Møller, B.L.; Meisel, L.; Oraguzie, N.; Silva, H.; et al. Comparative Genomics Analysis in Prunoideae to Identify Biologically Relevant Polymorphisms. Plant Biotechnol. J. 2013, 11, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Velasco, D.; Hough, J.; Aradhya, M.; Ross-Ibarra, J. Evolutionary Genomics of Peach and Almond Domestication. G3 Genes Genomes Genet. 2016, 6, 3985–3993. [Google Scholar] [CrossRef]
- Pérez de los Cobos, F.; Martínez-García, P.J.; Romero, A.; Miarnau, X.; Eduardo, I.; Howad, W.; Mnejja, M.; Dicenta, F.; Socias i Company, R.; Rubio-Cabetas, M.J.; et al. Pedigree Analysis of 220 Almond Genotypes Reveals Two World Mainstream Breeding Lines Based on Only Three Different Cultivars. Hortic. Res. 2021, 8, 11. [Google Scholar] [CrossRef]
- Krueger, F.; James, F.; Ewels, P.; Afyounian, E.; Schuster-Boeckler, B. FelixKrueger/TrimGalore: V0.6.7. Available online: https://github.com/FelixKrueger/TrimGalore/tree/0.6.7 (accessed on 23 July 2021).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 October 2022).
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinforma. Oxf. Engl. 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinforma. Oxf. Engl. 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinforma. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Pavan, S.; Delvento, C.; Ricciardi, L.; Lotti, C.; Ciani, E.; D’Agostino, N. Recommendations for Choosing the Genotyping Method and Best Practices for Quality Control in Crop Genome-Wide Association Studies. Front. Genet. 2020, 11, 447. [Google Scholar] [CrossRef]
- Dicenta, F.; Cremades, T.; Jose Martinez-Garcia, P.; Martinez-Gomez, P.; Ortega, E.; Rubio, M.; Sanchez-Perez, R.; Lopez-Alcolea, J.; Egea, J. Penta and Makako: Two Extra-Late Flowering Self-Compatible Almond Cultivars from CEBAS-CSIC. Hortscience 2018, 53, 1700–1702. [Google Scholar] [CrossRef]
- Antanaviciute, L.; Harrison, N.; Battey, N.H.; Harrison, R.J. An Inexpensive and Rapid Genomic DNA Extraction Protocol for Rosaceous Species. J. Hortic. Sci. Biotechnol. 2015, 90, 427–432. [Google Scholar] [CrossRef]


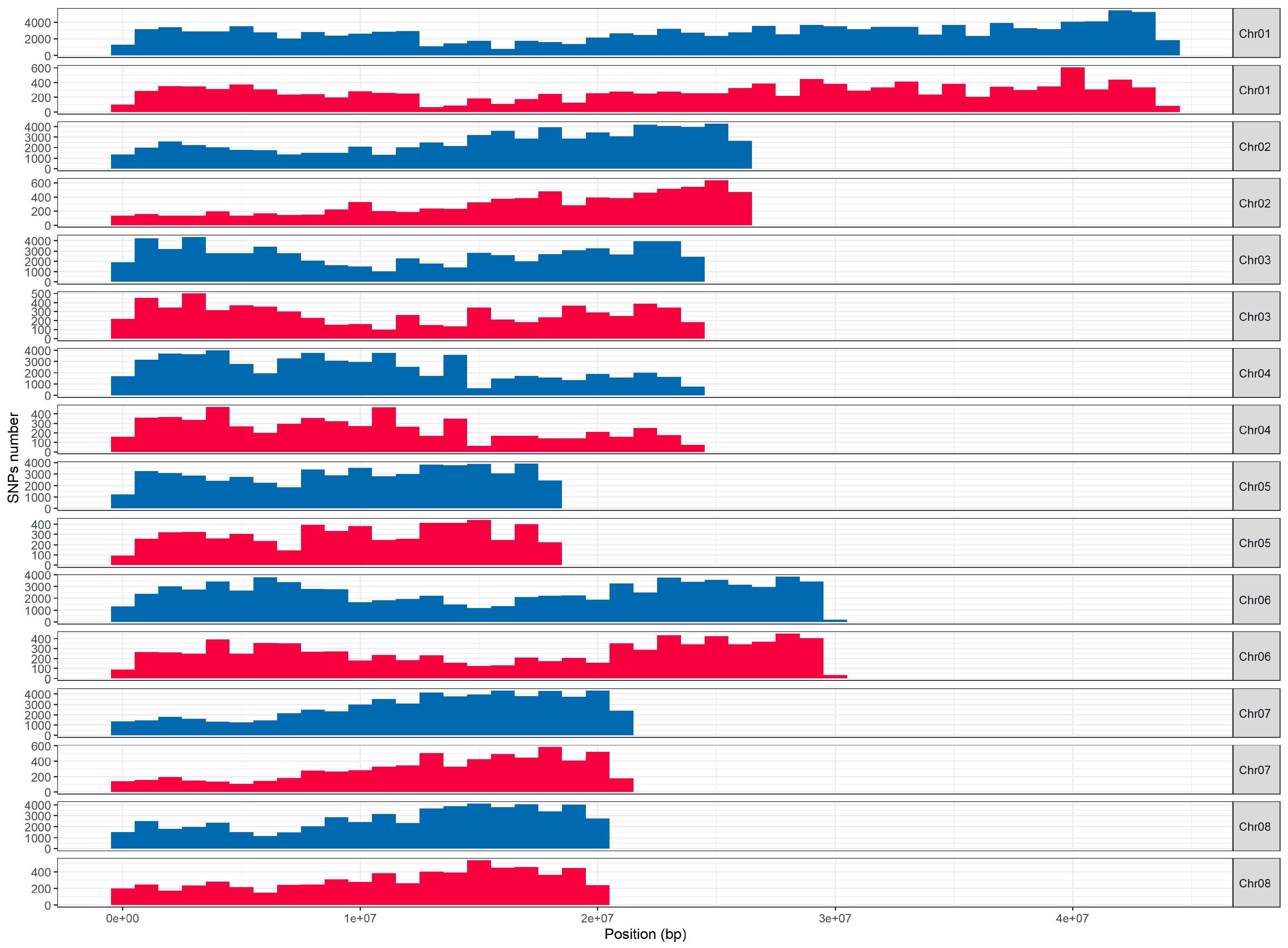
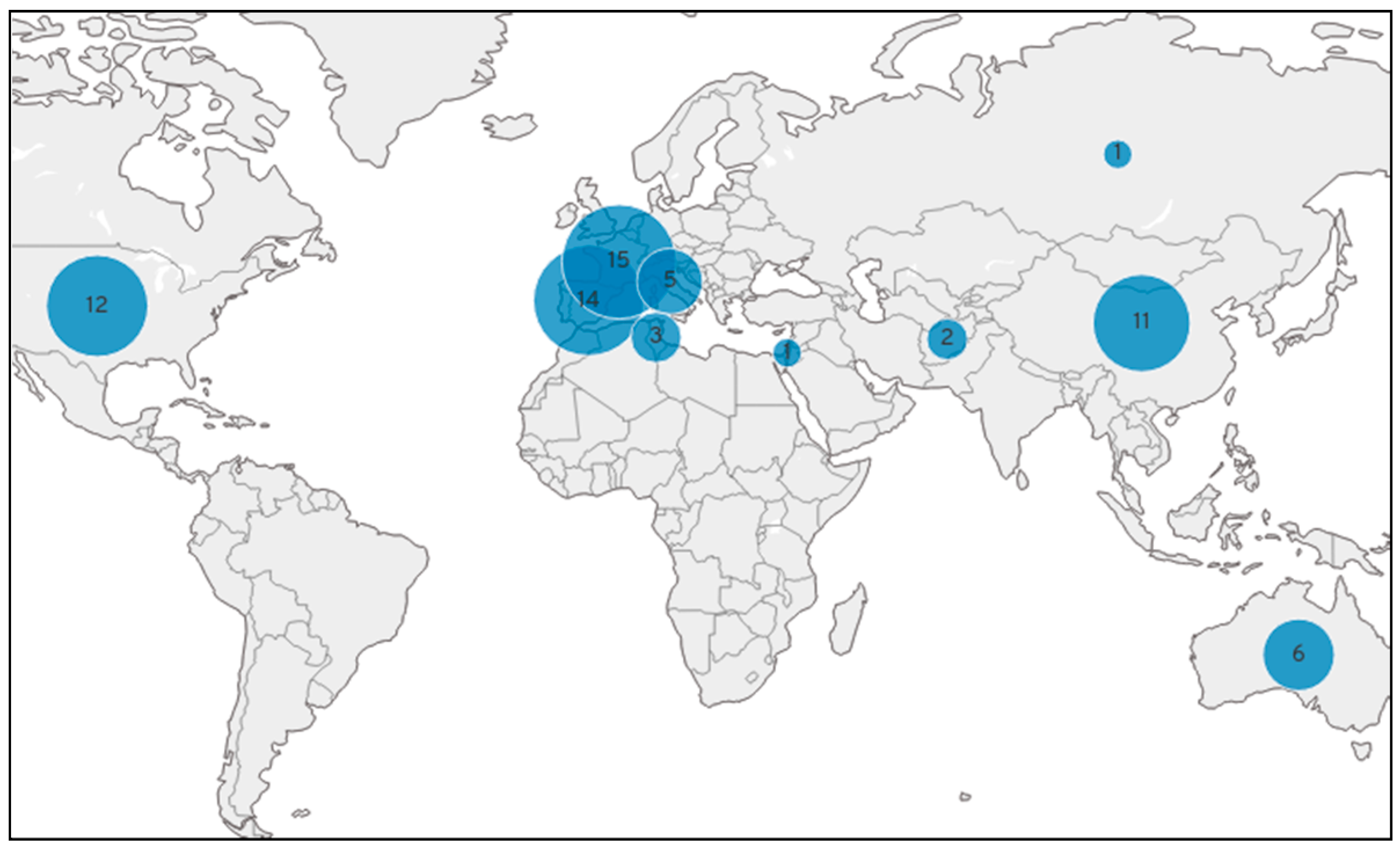
| SNP Name | Chr | Position_V2 | Position_V1 | SNP_Ref | SNP_Alt | Trait | Gene |
|---|---|---|---|---|---|---|---|
| AX-599403222 | Pd07 | 9,019,871 | 7,763,091 | A | G | RK Nematode R | RMja |
| AX-599403226 | Pd01 | 3,296,885 | 2,496,687 | T | C | Kernel weight | |
| AX-599403227 | Pd05 | 11,867,647 | 11,521,869 | C | T | Bitter taste | Sk |
| SNP Category | Number | Percent |
|---|---|---|
| Poly High Resolution (PHR) | 47,012 | 77.6 |
| Mono High Resolution (MHR) | 85 | 0.1 |
| No Minor Hom (NMH) | 3074 | 5.1 |
| Call Rate Below Threshold (CBT) | 2435 | 4 |
| Off-Target Variant (OTV) | 3425 | 5.7 |
| Other | 4550 | 7.5 |
| Total | 60,581 |
| Species | Accessions | NA Number | NA Percent | He Number | He Percent |
|---|---|---|---|---|---|
| P. dulcis | 185 * | 262–1665 | 0.4–2.7 | 7077–14,065 | 11.9–23.4 |
| P. bucharica | 1 | 3099 | 5 | 3242 | 6 |
| P. fenzliana | 9 | 1448–1937 | 2.4–3.2 | 3516–4267 | 5.9–7.3 |
| P. kuramica | 1 | 4212 | 7 | 3599 | 6 |
| P. dulcis × P. dehiscens | 2 | 576–1208 | 1–2 | 10,126–11,792 | 17.1–19.7 |
| P. dulcis × P. webbii | 1 | 570 | 1 | 14,976 | 25 |
| P. persica | 2 | 7103–9412 | 11.7–15.5 | 4965–5897 | 9.3–11.5 |
| P. davidiana | 2 | 5610–5671 | 9.3–9.4 | 4598–4822 | 8.4–8.8 |
| P. persica × P. dulcis | 3 | 1240–2003 | 2–3.3 | 6928–11,277 | 11.8–19 |
| P. bucharica × P. persica | 1 | 2130 | 4 | 4643 | 8 |
| P. pedunculata | 1 | 12,806 | 21 | 6809 | 14 |
| SNP Nb on “Penta” | Map | |||
|---|---|---|---|---|
| LG | He | p-Value (χ²) ≤ 0.01 | Marker Nb | Size (in cM) |
| 1 | 2055 | 1541 | 261 | 208 |
| 2 | 1339 | 534 | 81 | 69 |
| 3 | 1146 | 1146 | 166 | 108 |
| 4 | 1302 | 574 | 96 | 89 |
| 5 | 1042 | 451 | 87 | 96 |
| 6 | 1456 | 1402 | 190 | 114 |
| 7 | 1176 | 841 | 145 | 99 |
| 8 | 1158 | 745 | 133 | 94 |
| Total | 10,674 | 7234 | 1159 | 876 |
| Allele State * | Number | Percent |
|---|---|---|
| 0 | 109 | 0.2 |
| 1 | 10,416 | 17.2 |
| 2 | 49,765 | 82.1 |
| NA | 291 | 0.5 |
| Total | 60,581 |
| Replicate | R2 | R3 |
|---|---|---|
| R1 | 549 (0.90%) | 576 (0.95%) |
| R2 | - | 615 (1.01%) |
| F1 (“Penta”) | F2 Progeny | Erroneous Data Points | |||||
|---|---|---|---|---|---|---|---|
| Marker Nb | AA | AB | BB | Total | Nb | % | |
| AA | 20,462 | 999,542 | 494 | 2167 | 1,002,203 | 2661 | 0.27 |
| BB | 22,276 | 1324 | 332 | 1,089,549 | 1,091,205 | 1656 | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duval, H.; Coindre, E.; Ramos-Onsins, S.E.; Alexiou, K.G.; Rubio-Cabetas, M.J.; Martínez-García, P.J.; Wirthensohn, M.; Dhingra, A.; Samarina, A.; Arús, P. Development and Evaluation of an AxiomTM 60K SNP Array for Almond (Prunus dulcis). Plants 2023, 12, 242. https://doi.org/10.3390/plants12020242
Duval H, Coindre E, Ramos-Onsins SE, Alexiou KG, Rubio-Cabetas MJ, Martínez-García PJ, Wirthensohn M, Dhingra A, Samarina A, Arús P. Development and Evaluation of an AxiomTM 60K SNP Array for Almond (Prunus dulcis). Plants. 2023; 12(2):242. https://doi.org/10.3390/plants12020242
Chicago/Turabian StyleDuval, Henri, Eva Coindre, Sebastian E. Ramos-Onsins, Konstantinos G. Alexiou, Maria J. Rubio-Cabetas, Pedro J. Martínez-García, Michelle Wirthensohn, Amit Dhingra, Anna Samarina, and Pere Arús. 2023. "Development and Evaluation of an AxiomTM 60K SNP Array for Almond (Prunus dulcis)" Plants 12, no. 2: 242. https://doi.org/10.3390/plants12020242
APA StyleDuval, H., Coindre, E., Ramos-Onsins, S. E., Alexiou, K. G., Rubio-Cabetas, M. J., Martínez-García, P. J., Wirthensohn, M., Dhingra, A., Samarina, A., & Arús, P. (2023). Development and Evaluation of an AxiomTM 60K SNP Array for Almond (Prunus dulcis). Plants, 12(2), 242. https://doi.org/10.3390/plants12020242








