Characterization and Transcriptome Analysis of Maize Small-Kernel Mutant smk7a in Different Development Stages
Abstract
1. Introduction
2. Results
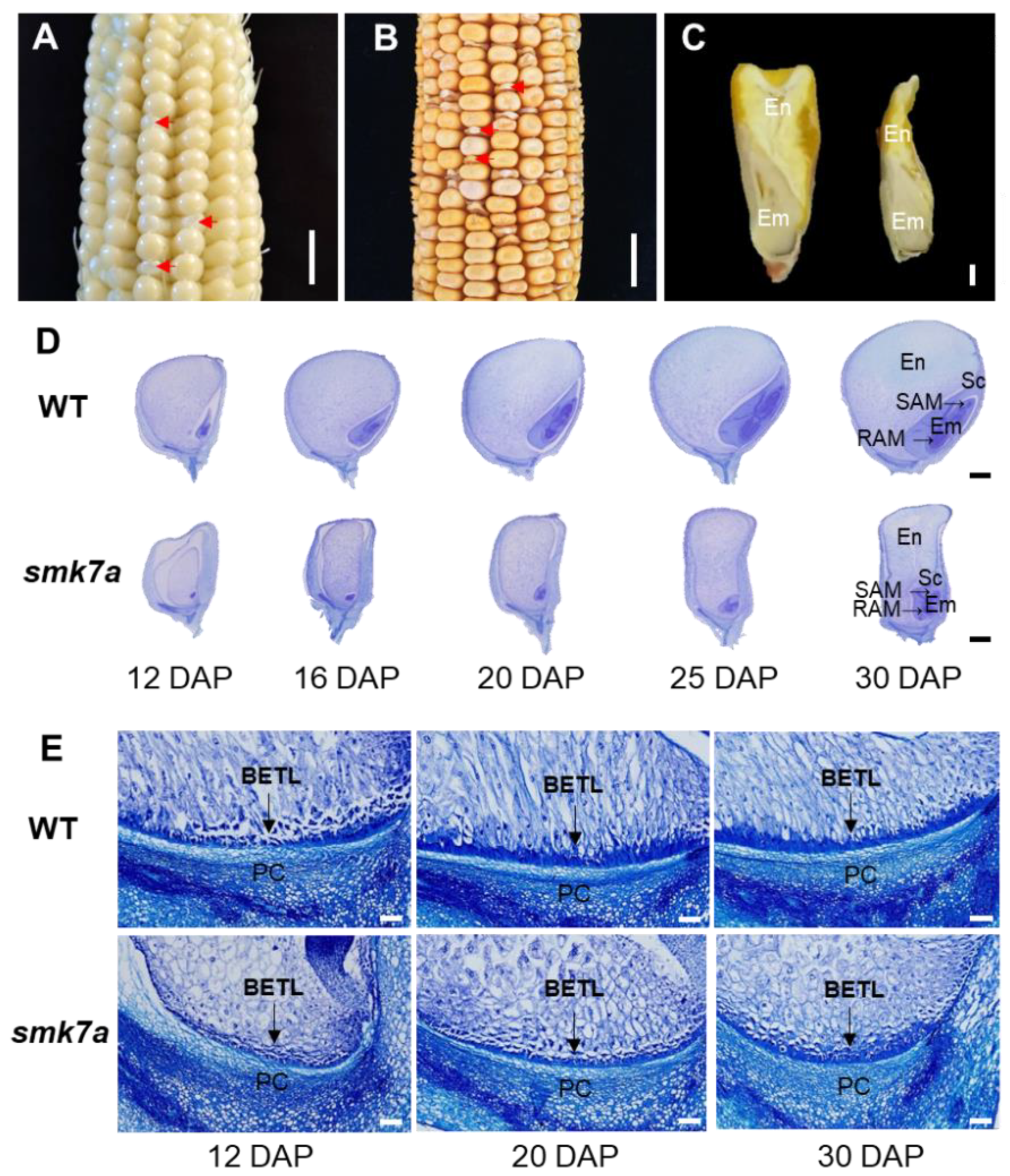
2.1. smk7a Mutant Exhibited Defective Embryo and Endosperm Development
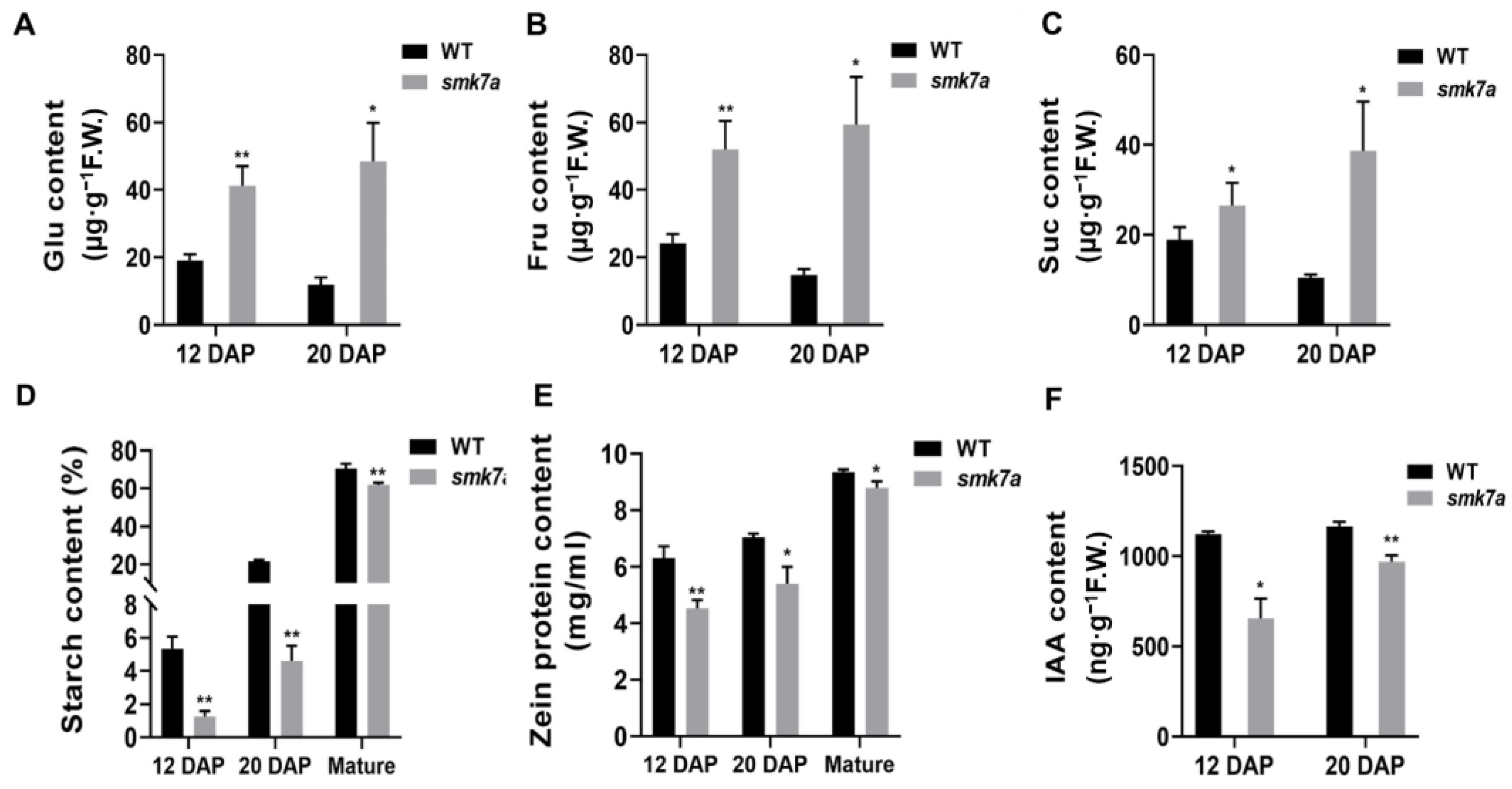
2.2. smk7a Mutant Had Lower Starch, Storage Protein, and Free IAA Contents
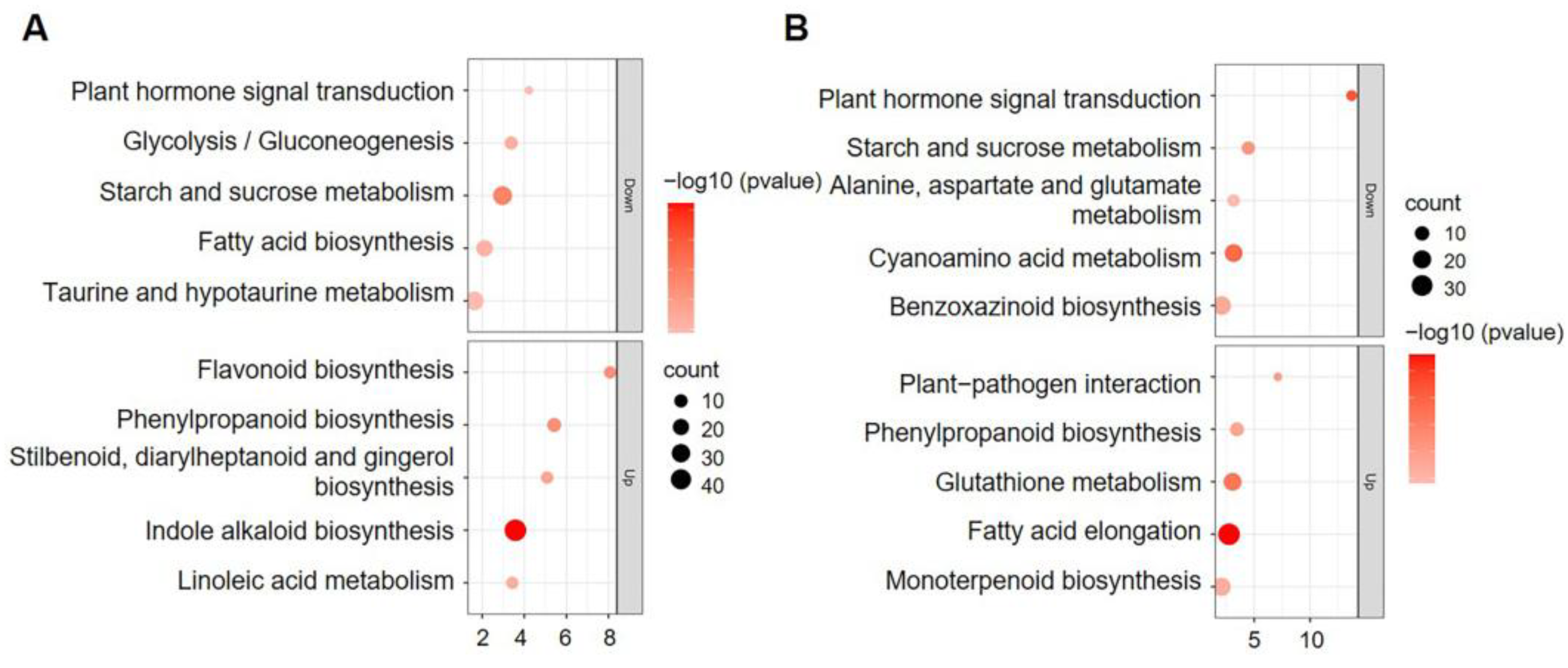
2.3. Transcriptome Comparison of Developing Kernels from WT and smk7a
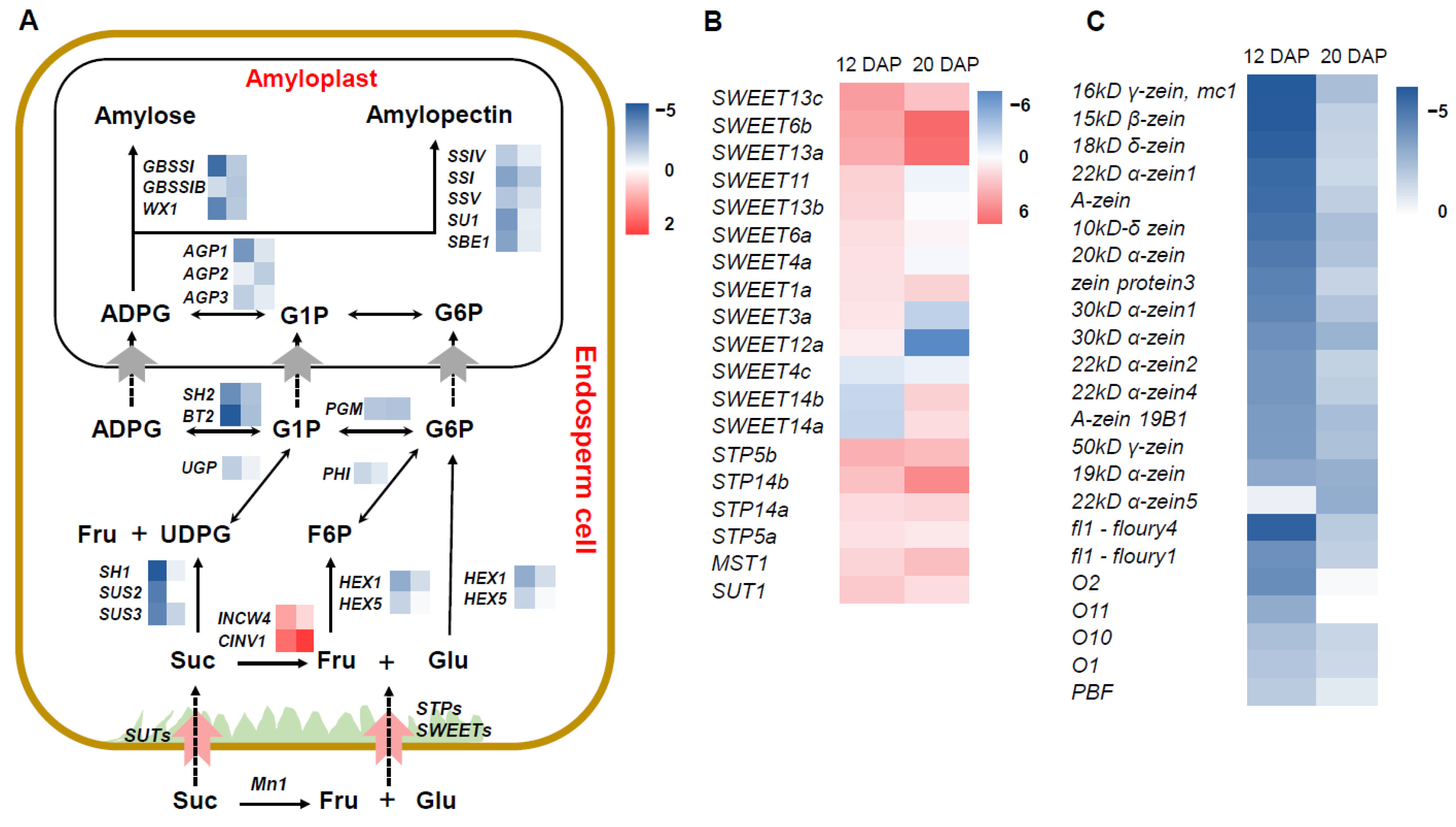
2.4. Suppressed Expression of Genes Involved in Starch Synthesis and Protein Storage in smk7a
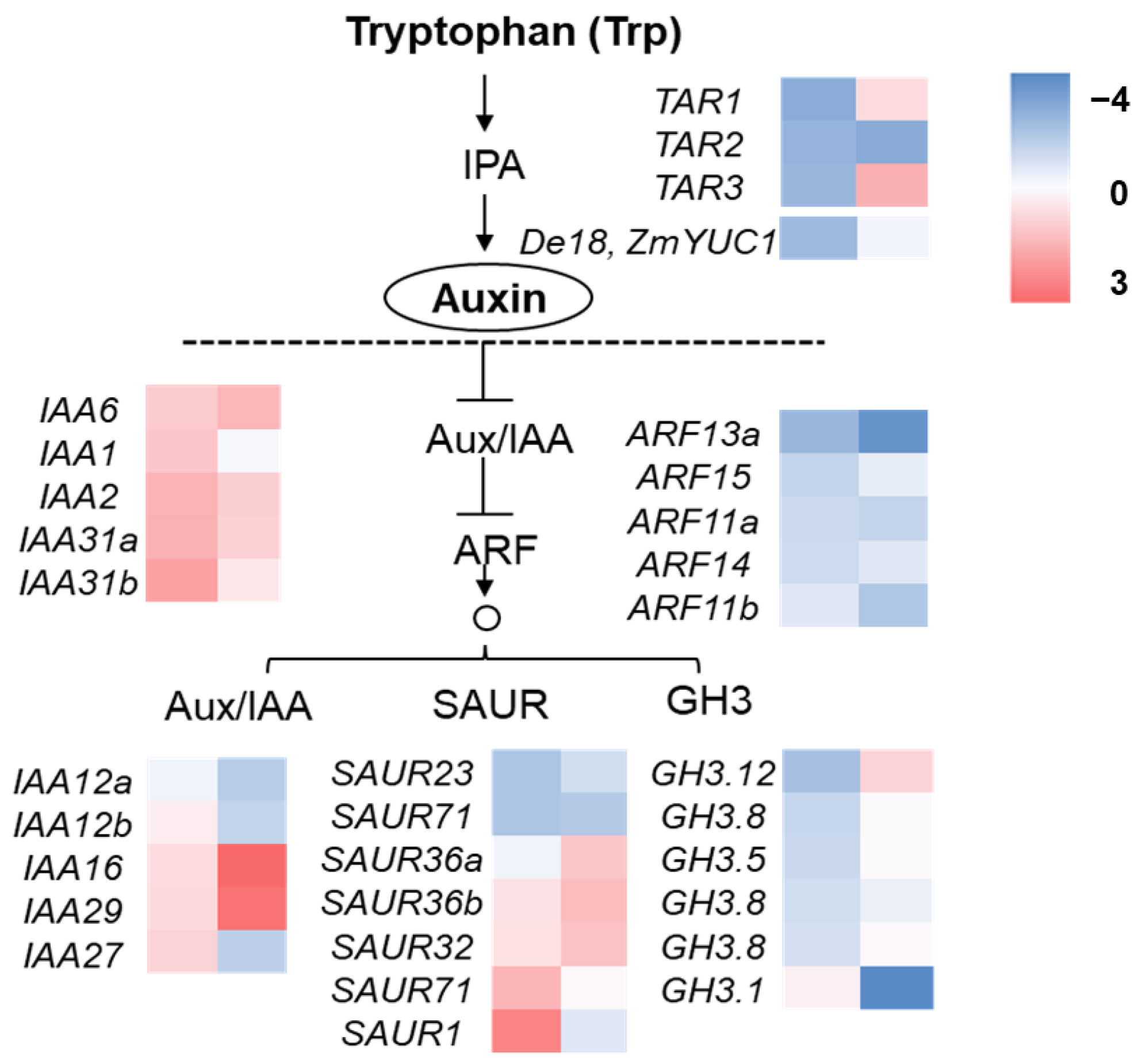
2.5. Disrupted Expression of Genes Related to IAA Synthesis and Signaling in smk7a
2.6. Genetic Analysis and Fine Mapping of the smk7a Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Light Microscopy and Scanning Electron Microscopy Observations of smk7a and WT Kernels
4.3. Measurement of Protein and Starch Contents
4.4. Measurement of Soluble Sugar and Free IAA Contents
4.5. RNA Sequencing Analysis
4.6. RNA Extraction and Quantitative RT-PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doll, N.M.; Depège-Fargeix, N.; Rogowsky, P.M.; Widiez, T. Signaling in Early Maize Kernel Development. Mol. Plant 2017, 10, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Royo, J.; Muñiz, L.M.; Sellam, O.; Paul, W.; Gerentes, D.; Barrero, C.; Ópez, M.; Perez, P.; Hueros, G. The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell 2009, 21, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.-Y.; Dong, Y.-B.; Lu, S.-P.; Liu, T.-S.; He, C.-M.; Liu, C.-X.; Liu, Q.; Dong, R.; Wang, J.; Li, Y.-L.; et al. Characterization and map-based cloning of miniature2-m1, a gene controlling kernel size in maize. J. Integr. Agric. 2020, 19, 1961–1973. [Google Scholar] [CrossRef]
- Hu, M.; Zhao, H.; Yang, B.; Yang, S.; Liu, H.; Tian, H.; Shui, G.; Chen, Z.; E, L.; Lai, J.; et al. ZmCTLP1 is required for the maintenance of lipid homeostasis and the basal endosperm transfer layer in maize kernels. New Phytol. 2021, 232, 2384–2399. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhong, M.; Shuai, B.; Song, J.; Zhang, J.; Han, L.; Ling, H.; Tang, Y.; Wang, G.; Song, R. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 2017, 214, 1563–1578. [Google Scholar] [CrossRef]
- Zheng, Y. Molecular mechanisms of maize endosperm transfer cell development. Plant Cell Rep. 2022, 41, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Ma, Z.; Song, R. Maize endosperm development. J. Integr. Plant Biol. 2021, 63, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Ma, Z.; Song, R. Maize kernel development. Mol. Breed. 2021, 41, 2. [Google Scholar] [CrossRef]
- Ma, Z.; Dooner, H.K. A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J. 2004, 37, 92–103. [Google Scholar] [CrossRef]
- Magnard, J.L.; Heckel, T.; Massonneau, A.; Wisniewski, J.P.; Cordelier, S.; Lassagne, H.; Perez, P.; Dumas, C.; Rogowsky, P.M. Morphogenesis of maize embryos requires ZmPRPL35-1 encoding a plastid ribosomal protein. Plant Physiol. 2004, 134, 649–663. [Google Scholar] [CrossRef]
- Li, C.; Shen, Y.; Meeley, R.; McCarty, D.R.; Tan, B.C. Embryo defective 14 encodes a plastid-targeted cGTPase essential for embryogenesis in maize. Plant J. 2015, 84, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, C.; McCarty, D.R.; Meeley, R.; Tan, B.C. Embryo defective12 encodes the plastid initiation factor 3 and is essential for embryogenesis in maize. Plant J. 2013, 74, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Hou, M.M.; Tan, B.C. The Requirement of WHIRLY1 for Embryogenesis Is Dependent on Genetic Background in Maize. PLoS ONE 2013, 8, e67369. [Google Scholar] [CrossRef] [PubMed]
- Becraft, P.W.; Stinard, P.S.; McCarty, D.R. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 1996, 273, 1406–1409. [Google Scholar] [CrossRef]
- Becraft, P.W.; Li, K.; Dey, N.; Asuncion-Crabb, Y. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 2002, 129, 5217–5225. [Google Scholar] [CrossRef]
- Lid, S.E.; Gruis, D.; Jung, R.; Lorentzen, J.A.; Ananiev, E.; Chamberlin, M.; Niu, X.M.; Meeley, R.; Nichols, S.; Olsen, O.A. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 2002, 99, 5460–5465. [Google Scholar] [CrossRef]
- Shen, B.; Li, C.; Min, Z.; Meeley, R.B.; Tarczynski, M.C.; Olsen, O.A. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc. Natl. Acad. Sci. USA 2003, 100, 6552–6557. [Google Scholar] [CrossRef]
- Yi, G.; Lauter, A.M.; Scott, M.P.; Becraft, P.W. The thick aleurone1 Mutant Defines a Negative Regulation of Maize Aleurone Cell Fate That Functions Downstream of defective kernel 1. Plant Physiol. 2011, 156, 1826–1836. [Google Scholar] [CrossRef]
- Wu, H.; Gontarek, B.C.; Yi, G.; Beall, B.D.; Neelakandan, A.K.; Adhikari, B.; Chen, R.; McCarty, D.R.; Severin, A.J.; Becraft, P.W. The thick aleurone1 Gene Encodes a NOT1 Subunit of the CCR4-NOT Complex and Regulates Cell Patterning in Endosperm. Plant Physiol. 2020, 184, 960–972. [Google Scholar] [CrossRef]
- Kang, B.-H.; Xiong, Y.; Williams, D.S.; Pozueta-Romero, D.; Chourey, P.S. Miniature1-Encoded Cell Wall Invertase Is Essential for Assembly and Function of Wall-in-Growth in the Maize Endosperm Transfer Cell. Plant Physiol. 2009, 151, 1366–1376. [Google Scholar] [CrossRef]
- LeCLere, S.; Schmelz, E.; Chourey, P. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010, 153, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, M.; Chen, J.; Qi, W.; Lai, J.; Ma, Z.; Song, R. ENB1 encodes a cellulose synthase 5 that directs synthesis of cell wall ingrowths in maize basal endosperm transfer cells. Plant Cell 2022, 34, 1054–1074. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Ópez-Coria, M.; Sánchez-Sánchez, T.; Martínez-Marcelo, V.; Aguilera-Alvarado, G.; Flores-Barrera, M.; King-Díaz, B.; Sánchez-Nieto, S. SWEET Transporters for the Nourishment of Embryonic Tissues during Maize Germination. Genes 2019, 10, 780. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.; Yu, M.; Zhang, M.; Zhong, Y.; Wang, T.; Liu, P.; Song, W.; Zhao, H.; Fastner, A.; et al. The sugar transporter ZmSUGCAR1 of the Nitrate Transporter 1/Peptide Transporter family is critical for maize grain filling. Plant Cell 2022, 34, 4232–4254. [Google Scholar] [CrossRef] [PubMed]
- Muñiz, L.M.; Royo, J.; Gómez, E.; Barrero, C.; Bergareche, D.; Hueros, G. The maize transfer cell-specific type-A response regulator ZmTCRR-1 appears to be involved in intercellular signalling. Plant J. 2006, 48, 17–27. [Google Scholar] [CrossRef]
- Muñiz, L.M.; Royo, J.; Gómez, E.; Baudot, G.; Paul, W.; Hueros, G. Atypical response regulators expressed in the maize endosperm transfer cells link canonical two component systems and seed biology. BMC Plant Biol. 2010, 10, 84. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20. [Google Scholar] [CrossRef]
- Finegan, C.; Boehlein, S.K.; Leach, K.A.; Madrid, G.; Hannah, L.C.; Koch, K.E.; Tracy, W.F.; Resende, M.F.R., Jr. Genetic Perturbation of the Starch Biosynthesis in Maize Endosperm Reveals Sugar-Responsive Gene Networks. Front. Plant Sci. 2021, 12, 800326. [Google Scholar] [CrossRef]
- Bonello, J.-F.; Opsahl-Ferstad, H.G.; Perez, P.; Dumas, C.; Rogowsky, P.M. Esr genes show different levels of expression in the same region of maize endosperm. Gene 2000, 246, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Tan, H.; Zhang, C.; Li, Q.; Liu, Q. Starch biosynthesis in cereal endosperms: An updated review over the last decade. Plant Commun. 2021, 2, 100237. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.G.; Shi, H.M.; Wang, H.W.; Li, K.; Huang, C.L.; Liu, Z.F.; Wu, Y.J.; Li, S.Q.; Hu, X.J.; Ma, Q. Phenotype analysis and gene mapping of small kernel 7 (smk7) mutant in maize. Acta Agron. Sin. 2021, 47, 285–293. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Z. Protein accumulation in aleurone cells, sub-aleurone cells and the center starch endosperm of cereals. Plant Cell Rep. 2014, 33, 1607–1615. [Google Scholar] [CrossRef]
- Leroux, B.M.; Goodyke, A.J.; Schumacher, K.I.; Abbott, C.P.; Clore, A.M.; Yadegari, R.; Larkins, B.A.; Dannenhoffer, J.M. Maize early endosperm growth and development: From fertilization through cell type differentiation. Am. J. Bot. 2014, 101, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Qi, W.; Song, R. Maize Dek15 Encodes the Cohesin-Loading Complex Subunit SCC4 and Is Essential for Chromosome Segregation and Kernel Development. Plant Cell 2019, 31, 465–485. [Google Scholar] [CrossRef]
- Wang, H.C.; Sayyed, A.; Liu, X.-Y.; Yang, Y.Z.; Sun, F.; Wang, Y.; Wang, M.; Tan, B.C. SMALL KERNEL4 is required for mitochondrial cox1 transcript editing and seed development in maize. J. Integr. Plant Biol. 2019, 62, 777–792. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, Y.F.; Hou, M.; Sun, F.; Shen, Y.; Xiu, Z.H.; Wang, X.; Chen, Z.L.; Sun, S.S.; Small, I.; et al. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 2014, 79, 797–809. [Google Scholar] [CrossRef]
- Li, P.; Li, Z.; Xie, G.; Zhang, J. ZmThx20Trihelix Transcription Factor Is Required for Kernel Development in Maize. Int. J. Mol. Sci. 2021, 22, 12137. [Google Scholar] [CrossRef]
- Shen, S.; Ma, S.; Chen, X.M.; Yi, F.; Li, B.B.; Liang, X.G.; Liao, S.J.; Gao, L.H.; Zhou, S.L.; Ruan, Y.L. A transcriptional landscape underlying sugar import for grain set in maize. Plant J. 2022, 110, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Almagro, G.; Baroja-Fernández, E.; Muñoz, F.J.; Bahaji, A.; Etxeberria, E.; Li, J.; Montero, M.; Hidalgo, M.; Sesma, M.T.; Pozueta-Romero, J. No evidence for the occurrence of substrate inhibition of Arabidopsis thaliana sucrose synthase-1 (AtSUS1) by fructose and UDP-glucose. Plant Signal. Behav. 2012, 7, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Baroja-Fernández, E.; Bahaji, A.; Muñoz, F.J.; Ovecka, M.; Montero, M.; Sesma, M.T.; Alonso-Casajús, N.; Almagro, G.; Sánchez-López, A.M.; et al. Enhancing sucrose synthase activity results in increased levels of starch and ADP-glucose in maize (Zea mays L.) seed endosperms. Plant Cell Physiol. 2013, 54, 282–294. [Google Scholar] [CrossRef]
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-Glucose Pyrophosphorylase: A Regulatory Enzyme for Plant Starch Synthesis. Photosynth. Res. 2004, 79, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, Y.; Weng, J.; Liu, H.; Yu, G.; Liu, Y.; Xiao, Q.; Huang, H.; Wang, Y.; Wei, B.; et al. Coordinated regulation of starch synthesis in maize endosperm by microRNAs and DNA methylation. Plant J. 2021, 105, 108–123. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, L.; Cheng, W.; Liu, B.; Li, W.; Wang, F.; Xu, C.; Zhao, X.; Ding, Z.; Zhang, K.; et al. SH1-dependent maize seed development and starch synthesis via modulating carbohydrate flow and osmotic potential balance. BMC Plant Biol. 2020, 20, 264. [Google Scholar] [CrossRef]
- Giroux, M.J.; Hannah, L.C. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Mol. Gen. Genet. 1994, 243, 400–408. [Google Scholar] [CrossRef]
- Huang, B.; Hennen-Bierwagen, T.A.; Myers, A.M. Functions of multiple genes encoding ADP-glucose pyrophosphorylase subunits in maize endosperm, embryo, and leaf. Plant Physiol. 2014, 164, 596–611. [Google Scholar] [CrossRef]
- Yu, G.; Lv, Y.; Shen, L.; Wang, Y.; Qing, Y.; Wu, N.; Li, Y.; Huang, H.; Zhang, N.; Liu, Y.; et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tag(tm) Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. Int. J. Mol. Sci. 2019, 20, 986. [Google Scholar]
- Zhan, J.; Li, G.; Ryu, C.H.; Ma, C.; Zhang, S.; Lloyd, A.; Hunter, B.G.; Larkins, B.A.; Drews, G.N.; Wang, X.; et al. Opaque-2 Regulates a Complex Gene Network Associated with Cell Differentiation and Storage Functions of Maize Endosperm. Plant Cell 2018, 30, 2425–2446. [Google Scholar] [CrossRef]
- Ning, L.; Wang, Y.; Shi, X.; Zhou, L.; Ge, M.; Liang, S.; Wu, Y.; Zhang, T.; Zhao, H. Nitrogen-dependent binding of the transcription factor PBF1 contributes to the balance of protein and carbohydrate storage in maize endosperm. Plant Cell 2022, 35, 409–434. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Qi, W.; Lv, Y.; Yan, S.; Xu, L.; Yang, W.; Yuan, Y.; Chen, Y.; Zhao, H.; Song, R. OPAQUE11 Is a Central Hub of the Regulatory Network for Maize Endosperm Development and Nutrient Metabolism. Plant Cell 2018, 30, 375–396. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, J.; Battaglia, R.; Bagnaresi, P.; Lucini, L.; Marocco, A. Transcriptomic and metabolomic analysis of ZmYUC1 mutant reveals the role of auxin during early endosperm formation in maize. Plant Sci. 2019, 281, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Silva-Sanchez, C.; Chen, S.; Li, J.; Chourey, P. A comparative glycoproteome study of developing endosperm in the hexose-deficient miniature1 (mn1) seed mutant and its wild type Mn1 in maize. Front. Plant Sci. 2014, 5, 63. [Google Scholar] [CrossRef]
- Chourey, P.S.; Li, Q.B.; Kumar, D. Sugar-hormone cross-talk in seed development: Two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol. Plant 2010, 3, 1026–1036. [Google Scholar] [CrossRef]
- Zhao, H.; Qin, Y.; Xiao, Z.; Li, Q.; Yang, N.; Pan, Z.; Gong, D.; Sun, Q.; Yang, F.; Zhang, Z.; et al. Loss of Function of an RNA Polymerase III Subunit Leads to Impaired Maize Kernel Development. Plant Physiol. 2020, 184, 359–373. [Google Scholar] [CrossRef]
- Murillo, M.M.S.; Granados-Chinchilla, F. Total starch in animal feeds and silages based on the chromatographic determination of glucose. MethodsX 2018, 5, 83–89. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.; Zhu, Y.; Huang, X.; Li, S.; Wu, X.; Zhao, Y.; Bao, Z.; Qin, L.; Jin, Y.; et al. THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 2022, 612, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.D.; Yin, J.; Hao, Y.H.; Li, Y.N.; Yuan, B.F.; Feng, Y.Q. Profiling of phytohormones in rice under elevated cadmium concentration levels by magnetic solid-phase extraction coupled with liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2015, 1406, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cai, W.J.; Wang, W.; Sun, M.X.; Feng, Y.Q. Rapid Analysis of Monosaccharides in Sub-milligram Plant Samples Using Liquid Chromatography-Mass Spectrometry Assisted by Post-column Derivatization. J. Agric. Food Chem. 2020, 68, 2588–2596. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
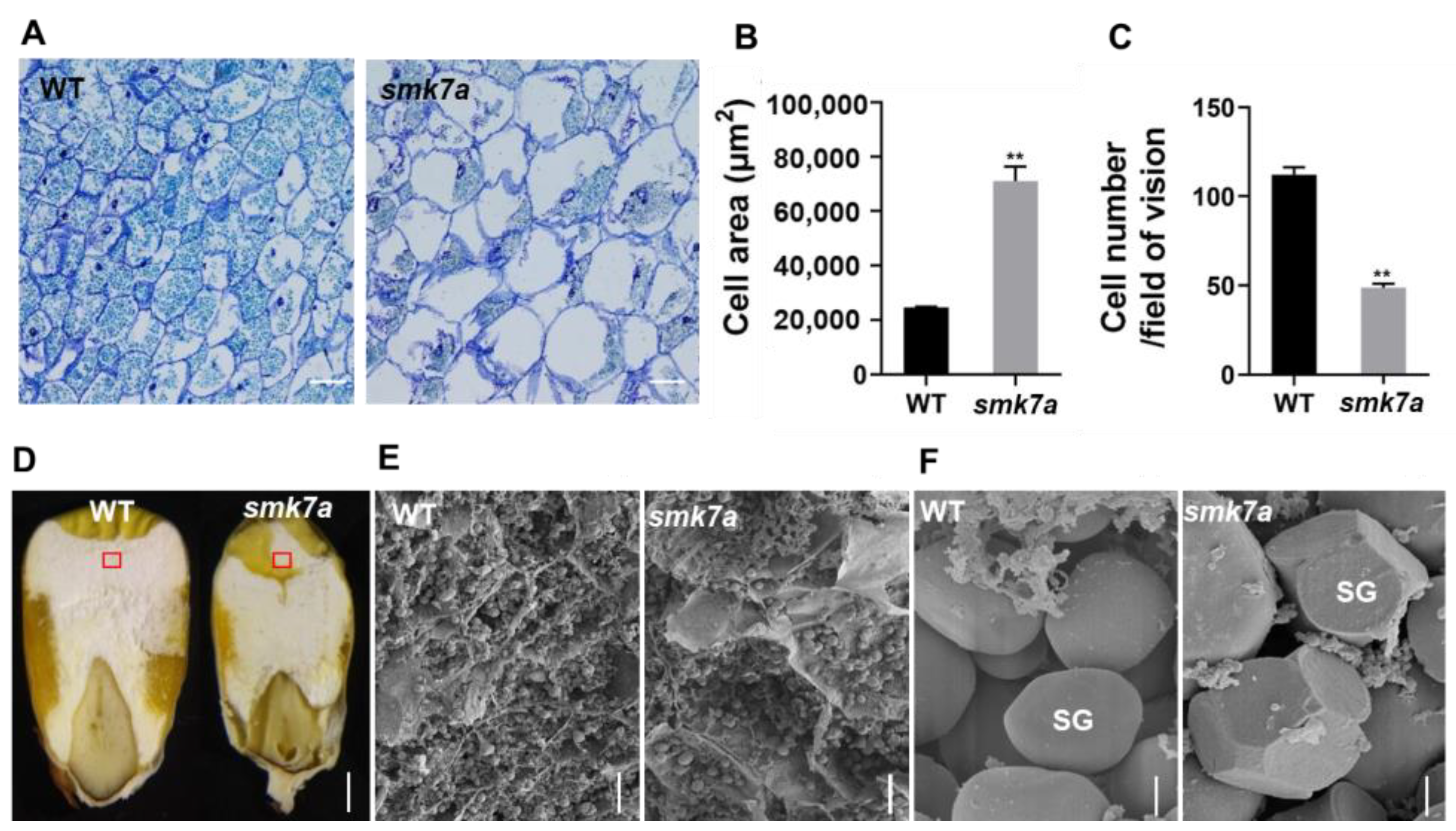
| WT | smk7a | Percentage Change | p Value | |
|---|---|---|---|---|
| Cell area (μm2) | 24,616.84 | 71,077.38 | 188.7% | 9.06 × 10−6 |
| Cell number | 112 | 49 | −56.3% | 2.02 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wang, H.; Li, K.; Liu, X.; Cao, X.; Zhou, Y.; Huang, C.; Peng, Y.; Hu, X. Characterization and Transcriptome Analysis of Maize Small-Kernel Mutant smk7a in Different Development Stages. Plants 2023, 12, 354. https://doi.org/10.3390/plants12020354
Wang J, Wang H, Li K, Liu X, Cao X, Zhou Y, Huang C, Peng Y, Hu X. Characterization and Transcriptome Analysis of Maize Small-Kernel Mutant smk7a in Different Development Stages. Plants. 2023; 12(2):354. https://doi.org/10.3390/plants12020354
Chicago/Turabian StyleWang, Jing, Hongwu Wang, Kun Li, Xiaogang Liu, Xiaoxiong Cao, Yuqiang Zhou, Changling Huang, Yunling Peng, and Xiaojiao Hu. 2023. "Characterization and Transcriptome Analysis of Maize Small-Kernel Mutant smk7a in Different Development Stages" Plants 12, no. 2: 354. https://doi.org/10.3390/plants12020354
APA StyleWang, J., Wang, H., Li, K., Liu, X., Cao, X., Zhou, Y., Huang, C., Peng, Y., & Hu, X. (2023). Characterization and Transcriptome Analysis of Maize Small-Kernel Mutant smk7a in Different Development Stages. Plants, 12(2), 354. https://doi.org/10.3390/plants12020354







