The Arabidopsis LHT1 Amino Acid Transporter Contributes to Pseudomonas simiae-Mediated Plant Growth Promotion by Modulating Bacterial Metabolism in the Rhizosphere
Abstract
1. Introduction
2. Results
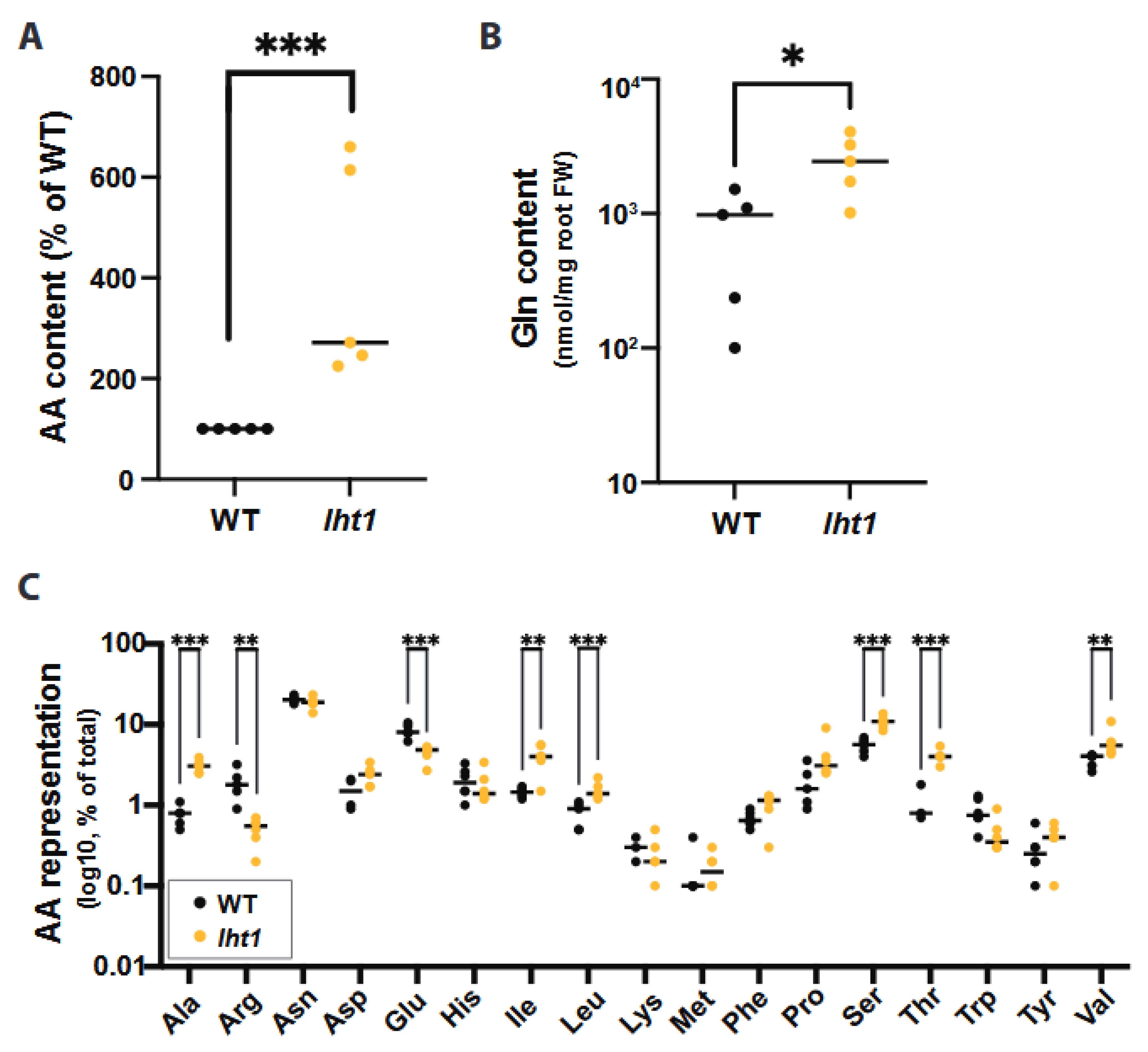
2.1. The Amino Acid Transporter LHT1 Modulates Amino Acid Content in Arabidopsis Root Exudates
2.2. The lht1 Root Exudates Enhance Ps WCS417r Growth
2.3. Loss of LHT1 Promotes Root Colonization by Ps WCS417r
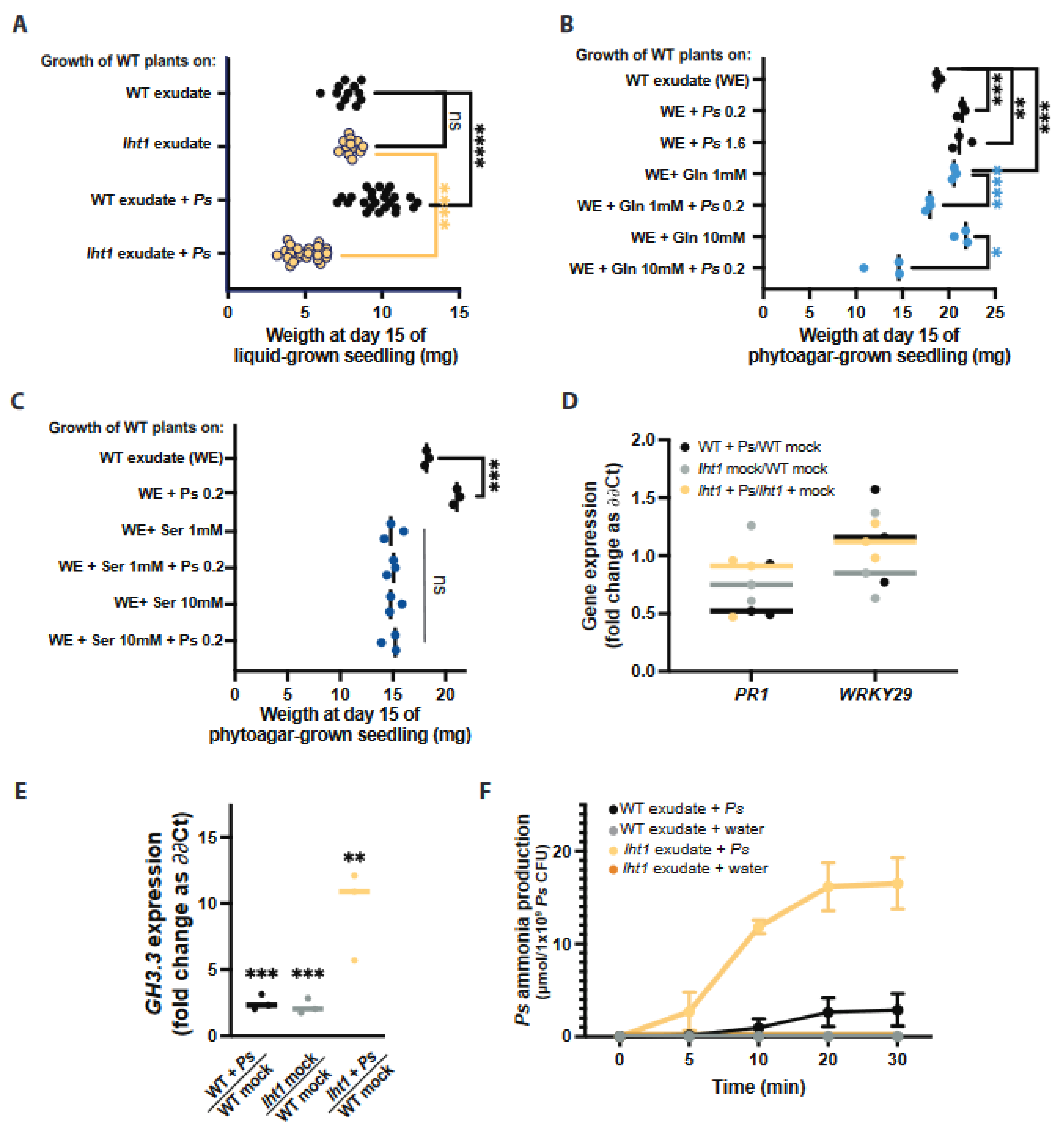
2.4. Rhizospheric Amino Acids Stimulate Robust Root Colonization but Compromise Plant Growth in a Microbial-Dependent Manner
3. Discussion
3.1. LHT1 Activity Is Essential to Maintain a Balanced Composition of Amino Acids in the Root Exudates
3.2. The Re-Uptake of Root-Exuded Amino Acids Is Necessary to Modulate Bacterial Metabolism and Promote Plant Growth
4. Conclusions
5. Methods
5.1. Plant Materials
5.2. Growth of Seedlings in Vertical Plates
5.3. Root Exudate Collection Assays
5.4. Colorimetric/Fluorometric Quantitation of Amino Acids
5.5. Liquid Chromatography–Mass Spectrometry Analysis of Amino Acids
5.6. Bacterial Growth on Root Exudates
5.7. Root Colonization
5.8. Competitive Chemotaxis Assay
5.9. Biofilm Formation Assays
5.10. Plant Growth on Exudates ± Ps WCS417r
5.11. Ps WCS417r Excreted Ammonium Assessment
5.12. Plant Tissue Gene Expression Analysis
5.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pieterse, C.M.J.; Berendsen, R.L.; de Jonge, R.; Stringlis, I.A.; Van Dijken, A.J.H.; Van Pelt, J.A.; Van Wees, S.C.M.; Yu, K.; Zamioudis, C.; Bakker, P.A.H.M. Pseudomonas Simiae WCS417: Star Track of a Model Beneficial Rhizobacterium. Plant Soil 2021, 461, 245–263. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Jaeger, C.H., III; Lindow, S.E.; Miller, W.; Clark, E. Mapping of Sugar and Amino Acid Availability in Soil around Roots with Bacterial Sensors of Sucrose and Tryptophan. Appl. Environ. Microbiol. 1999, 65, 2685. [Google Scholar] [CrossRef] [PubMed]
- Strehmel, N.; Böttcher, C.; Schmidt, S.; Scheel, D. Profiling of Secondary Metabolites in Root Exudates of Arabidopsis Thaliana. Phytochemistry 2014, 108, 35–46. [Google Scholar] [CrossRef]
- Defoirdt, T. Amino Acid–Derived Quorum Sensing Molecules Controlling the Virulence of Vibrios (and Beyond). PLoS Pathog. 2019, 15, e1007815. [Google Scholar] [CrossRef]
- Champalal, L.; Kumar, U.S.; Krishnan, N.; Vaseeharan, B.; Mariappanadar, V.; Raman, P. Modulation of Quorum Sensing-Controlled Virulence Factors in Chromobacterium violaceum by Selective Amino Acids. FEMS Microbiol. Lett. 2018, 365, fny252. [Google Scholar] [CrossRef]
- Wen, J.; Yu, Y.; Chen, M.; Cui, L.; Xia, Q.; Zeng, X.; Guo, Y.; Pan, D.; Wu, Z. Amino Acid-Derived Quorum Sensing Molecule Alanine on the Gastrointestinal Tract Tolerance of the Lactobacillus Strains in the Cocultured Fermentation Model. Microbiol. Spectr. 2022, 10, e00832-21. [Google Scholar] [CrossRef]
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of Chemotaxis Sensory Proteins for Amino Acids in Pseudomonas fluorescens Pf0-1 and Their Involvement in Chemotaxis to Tomato Root Exudate and Root Colonization. Microbes Environ. 2012, 27, 462–469. [Google Scholar] [CrossRef]
- Acosta Aragón, Y.; Jatkauskas, J.; Vrotniakienė, V. The Effect of a Silage Inoculant on Silage Quality, Aerobic Stability, and Meat Production on Farm Scale. ISRN Vet. Sci. 2012, 2012, 345927. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, L.; Zhang, N.; Li, Z.; Zhang, G.; Xu, Y.; Shen, Q.; Zhang, R. Plant-Microbe Communication Enhances Auxin Biosynthesis by a Root-Associated Bacterium, Bacillus amyloliquefaciens SQR9. Mol. Plant-Microbe Interact. 2016, 29, 324–330. [Google Scholar] [CrossRef]
- Liu, Y.; Von Wirén, N. Ammonium as a Signal for Physiological and Morphological Responses in Plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar] [CrossRef]
- Britto, D.T.; Siddiqi, M.Y.; Glass, A.D.M.; Kronzucker, H.J. Futile Transmembrane NH4+ Cycling: A Cellular Hypothesis to Explain Ammonium Toxicity in Plants. Proc. Natl. Acad. Sci. USA 2001, 98, 4255–4258. [Google Scholar] [CrossRef] [PubMed]
- Näsholm, T.; Ekblad, A.; Nordin, A.; Giesler, R.; Högberg, M.; Högberg, P. Boreal Forest Plants Take up Organic Nitrogen. Nature 1998, 392, 914–917. [Google Scholar] [CrossRef]
- Melin, E.; Nilsson, H. Transfer of Labelled Nitrogen from Glutamic Acid to Pine Seedlings through the Mycelium of Boletus variegatus (Sw.) Fr. Nature 1953, 171, 134. [Google Scholar] [CrossRef] [PubMed]
- Lipson, D.; Näsholm, T. The Unexpected Versatility of Plants: Organic Nitrogen Use and Availability in Terrestrial Ecosystems. Oecologia 2001, 128, 305–316. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, Q.; Kraus, T.E.C.; Dahlgren, R.A.; Anastasio, C.; Zasoski, R.J. Contribution of Amino Compounds to Dissolved Organic Nitrogen in Forest Soils. Biogeochemistry 2002, 61, 173–198. [Google Scholar] [CrossRef]
- Senwo, Z.N.; Tabatabai, M.A. Amino Acid Composition of Soil Organic Matter. Biol. Fertil. Soils 1998, 26, 235–242. [Google Scholar] [CrossRef]
- Wipf, D.; Ludewig, U.; Tegeder, M.; Rentsch, D.; Koch, W.; Frommer, W.B. Conservation of Amino Acid Transporters in Fungi, Plants and Animals. Trends Biochem. Sci. 2002, 27, 139–147. [Google Scholar] [CrossRef]
- Lalonde, S.; Wipf, D.; Frommer, W.B. Transport Mechanisms For Organic Forms Of Carbon And Nitrogen Between Source And Sink. Annu. Rev. Plant Biol. 2004, 55, 341–372. [Google Scholar] [CrossRef]
- Okumoto, S.; Pilot, G. Amino Acid Export in Plants: A Missing Link in Nitrogen Cycling. Mol. Plant 2011, 4, 453–463. [Google Scholar] [CrossRef]
- Hirner, A.; Ladwig, F.; Stransky, H.; Okumoto, S.; Keinath, M.; Harms, A.; Frommer, W.B.; Koch, W. Arabidopsis LHT1 Is a High-Affinity Transporter for Cellular Amino Acid Uptake in Both Root Epidermis and Leaf Mesophyll. Plant Cell 2006, 18, 1931–1946. [Google Scholar] [CrossRef]
- Svennerstam, H.; Ganeteg, U.; Bellini, C.; Näsholm, T. Comprehensive Screening of Arabidopsis Mutants Suggests the Lysine Histidine Transporter 1 to Be Involved in Plant Uptake of Amino Acids. Plant Physiol. 2007, 143, 1853–1860. [Google Scholar] [CrossRef]
- Svennerstam, H.; Ganeteg, U.; Näsholm, T. Root Uptake of Cationic Amino Acids by Arabidopsis Depends on Functional Expression of Amino Acid Permease 5. New Phytol. 2008, 180, 620–630. [Google Scholar] [CrossRef]
- Svennerstam, H.; Jämtgård, S.; Ahmad, I.; Huss-Danell, K.; Näsholm, T.; Ganeteg, U. Transporters in Arabidopsis Roots Mediating Uptake of Amino Acids at Naturally Occurring Concentrations. New Phytol. 2011, 191, 459–467. [Google Scholar] [CrossRef]
- Lehmann, S.; Gumy, C.; Blatter, E.; Boeffel, S.; Fricke, W.; Rentsch, D. In Planta Function of Compatible Solute Transporters of the AtProT Family. J. Exp. Bot. 2011, 62, 787–796. [Google Scholar] [CrossRef]
- Fischer, W.-N.; Kwart, M.; Hummel, S.; Frommer, W.B. Substrate Specificity and Expression Profile of Amino Acid Transporters (AAPs) in Arabidopsis. J. Biol. Chem. 1995, 270, 16315–16320. [Google Scholar] [CrossRef]
- Grallath, S.; Weimar, T.; Meyer, A.; Gumy, C.; Suter-Grotemeyer, M.; Neuhaus, J.-M.; Rentsch, D. The AtProT Family. Compatible Solute Transporters with Similar Substrate Specificity But Differential Expression Patterns. Plant Physiol. 2005, 137, 117–126. [Google Scholar] [CrossRef]
- Hida, A.; Oku, S.; Kawasaki, T.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of the McpA and McpM Genes, Encoding Methyl-Accepting Proteins Involved in Amino Acid and l-Malate Chemotaxis, and Involvement of McpM-Mediated Chemotaxis in Plant Infection by Ralstonia pseudosolanacearum (Formerly Ralstonia solanacearum Phylotypes I and III). Appl. Environ. Microbiol. 2015, 81, 7420–7430. [Google Scholar] [CrossRef]
- Webb, B.A.; Helm, R.F.; Scharf, B.E. Contribution of Individual Chemoreceptors to Sinorhizobium meliloti Chemotaxis Towards Amino Acids of Host and Nonhost Seed Exudates. Mol. Plant. Microbe Interact. 2016, 29, 231–239. [Google Scholar] [CrossRef]
- Moe, L.A. Amino Acids in the Rhizosphere: From Plants to Microbes. Am. J. Bot. 2013, 100, 1692–1705. [Google Scholar] [CrossRef]
- Haney, C.H.; Samuel, B.S.; Bush, J.; Ausubel, F.M. Associations with Rhizosphere Bacteria Can Confer an Adaptive Advantage to Plants. Nat. Plants 2015, 1, 15051. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Khadka, P.; Puchalski, P.; Leehan, J.D.; Rossi, F.R.; Okumoto, S.; Pilot, G.; Danna, C.H. MAMP-Elicited Changes in Amino Acid Transport Activity Contribute to Restricting Bacterial Growth. Plant Physiol. 2022, 189, 2315–2331. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Nie, J.; Bai, R.; Sui, X. Amino Acid Transporters in Plants: Identification and Function. Plants 2020, 9, 972. [Google Scholar] [CrossRef] [PubMed]
- Allard-Massicotte, R.; Tessier, L.; Lécuyer, F.; Lakshmanan, V.; Lucier, J.; Garneau, D.; Caudwell, L.; Vlamakis, H.; Bais, H.P.; Beauregard, P.B. Bacillus Subtilis Early Colonization of Arabidopsis Thaliana Roots Involves Multiple Chemotaxis Receptors. MBio 2016, 7, e01664-16. [Google Scholar] [CrossRef]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.M.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J.J. Flagella-Driven Chemotaxis towards Exudate Components Is an Important Trait for Tomato Root Colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, D.; Liu, Y.; Li, S.; Shen, Q.; Zhang, R. Effects of Different Plant Root Exudates and Their Organic Acid Components on Chemotaxis, Biofilm Formation and Colonization by Beneficial Rhizosphere-Associated Bacterial Strains. Plant Soil 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, N.; Huang, Q.; Raza, W.; Li, R.; Vivanco, J.M.; Shen, Q. Organic Acids from Root Exudates of Banana Help Root Colonization of PGPR Strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 2015, 5, 13438. [Google Scholar] [CrossRef]
- Liu, X.; Parales, R.E. Bacterial Chemotaxis to Atrazine and Related S-Triazines. Appl. Environ. Microbiol. 2009, 75, 5481–5488. [Google Scholar] [CrossRef]
- Heil, M.; Baldwin, I.T. Fitness Costs of Induced Resistance: Emerging Experimental Support for a Slippery Concept. Trends Plant Sci. 2002, 7, 61–67. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef]
- Burdon, J.J.; Thrall, P.H. The Fitness Costs to Plants of Resistance to Pathogens. Genome Biol. 2003, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Herms, D.A.; Mattson, W.J. The Dilemma of Plants: To Grow or Defend. Q. Rev. Biol. 1992, 67, 283–335. [Google Scholar] [CrossRef]
- Coley, P.D.; Bryant, J.P.; Chapin, F.S. Resource Availability and Plant Antiherbivore Defense. Science 1985, 230, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Maleck, K.; Levine, A.; Eulgem, T.; Morgan, A.; Schmid, J.; Lawton, K.A.; Dangl, J.L.; Dietrich, R.A. The Transcriptome of Arabidopsis Thaliana during Systemic Acquired Resistance. Nat. Genet. 2000, 26, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. Map Kinase Signalling Cascade in Arabidopsis Innate Immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-Mediated Auxin Homeostasis Links Growth Regulation with Stress Adaptation Response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Di, D.-W.; Li, G.; Sun, L.; Wu, J.; Wang, M.; Kronzucker, H.J.; Fang, S.; Chu, J.; Shi, W. High Ammonium Inhibits Root Growth in Arabidopsis Thaliana by Promoting Auxin Conjugation Rather than Inhibiting Auxin Biosynthesis. J. Plant Physiol. 2021, 261, 153415. [Google Scholar] [CrossRef]
- Savage, J.A.; Clearwater, M.J.; Haines, D.F.; Klein, T.; Mencuccini, M.; Sevanto, S.; Turgeon, R.; Zhang, C. Allocation, Stress Tolerance and Carbon Transport in Plants: How Does Phloem Physiology Affect Plant Ecology? Plant Cell Environ. 2016, 39, 709–725. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon Flow in the Rhizosphere: Carbon Trading at the Soil-Root Interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Proietti, S.; Hickman, R.; Van Verk, M.C.; Zamioudis, C.; Pieterse, C.M.J. Root Transcriptional Dynamics Induced by Beneficial Rhizobacteria and Microbial Immune Elicitors Reveal Signatures of Adaptation to Mutualists. Plant J. 2018, 417, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The Plant Microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of Action of Plant Growth Promoting Bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by Small-Molecule Hormones in Plant Immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Sharma, S.; Lin, W.; Villamor, J.G.; Verslues, P.E. Divergent Low Water Potential Response in Arabidopsis Thaliana Accessions Landsberg Erecta and Shahdara. Plant. Cell Environ. 2013, 36, 994–1008. [Google Scholar] [CrossRef]
- Fomsgaard, I.S.; Mortensen, A.G.; Carlsen, S.C.K. Microbial Transformation Products of Benzoxazolinone and Benzoxazinone Allelochemicals—A Review. Chemosphere 2004, 54, 1025–1038. [Google Scholar] [CrossRef]
- Soulas, G.; Lagacherie, B. Modelling of Microbial Degradation of Pesticides in Soils. Biol. Fertil. Soils 2001, 33, 551–557. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Nemkov, T.; D’Alessandro, A.; Hansen, K.C. Three-Minute Method for Amino Acid Analysis by UHPLC and High-Resolution Quadrupole Orbitrap Mass Spectrometry. Amino Acids 2015, 47, 2345–2357. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zerbe, G.O. Randomization Analysis of the Completely Randomized Design Extended to Growth and Response Curves. J. Am. Stat. Assoc. 1979, 74, 215–221. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agorsor, I.D.K.; Kagel, B.T.; Danna, C.H. The Arabidopsis LHT1 Amino Acid Transporter Contributes to Pseudomonas simiae-Mediated Plant Growth Promotion by Modulating Bacterial Metabolism in the Rhizosphere. Plants 2023, 12, 371. https://doi.org/10.3390/plants12020371
Agorsor IDK, Kagel BT, Danna CH. The Arabidopsis LHT1 Amino Acid Transporter Contributes to Pseudomonas simiae-Mediated Plant Growth Promotion by Modulating Bacterial Metabolism in the Rhizosphere. Plants. 2023; 12(2):371. https://doi.org/10.3390/plants12020371
Chicago/Turabian StyleAgorsor, Israel D. K., Brian T. Kagel, and Cristian H. Danna. 2023. "The Arabidopsis LHT1 Amino Acid Transporter Contributes to Pseudomonas simiae-Mediated Plant Growth Promotion by Modulating Bacterial Metabolism in the Rhizosphere" Plants 12, no. 2: 371. https://doi.org/10.3390/plants12020371
APA StyleAgorsor, I. D. K., Kagel, B. T., & Danna, C. H. (2023). The Arabidopsis LHT1 Amino Acid Transporter Contributes to Pseudomonas simiae-Mediated Plant Growth Promotion by Modulating Bacterial Metabolism in the Rhizosphere. Plants, 12(2), 371. https://doi.org/10.3390/plants12020371





