A Gene Encoding Xylanase Inhibitor Is a Candidate Gene for Bruchid (Callosobruchus spp.) Resistance in Zombi Pea (Vigna vexillata (L.) A. Rich)
Abstract
:1. Introduction
2. Results
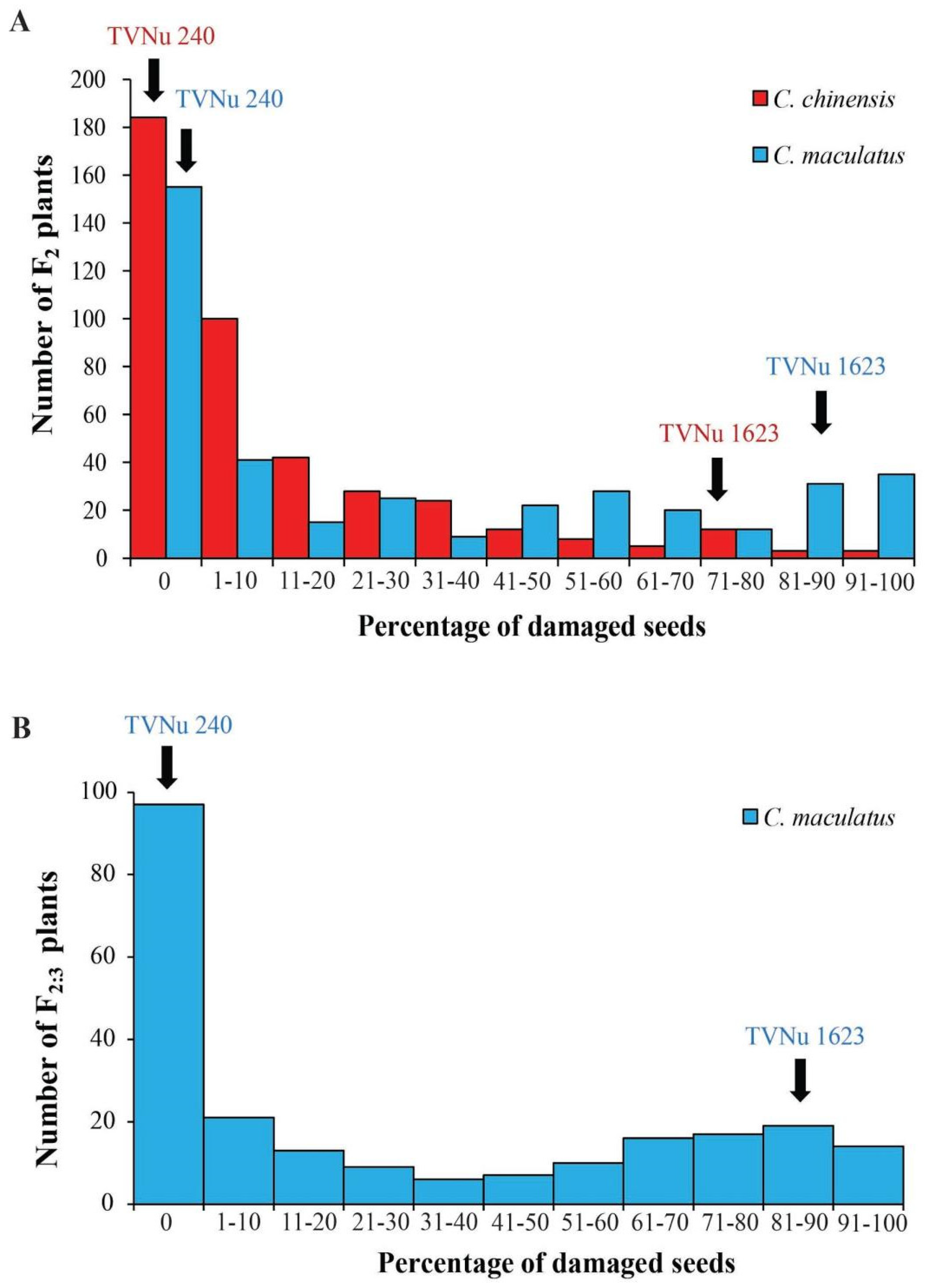
2.1. Variation in Resistance to C. maculatus and C. chinensis in the Parents and F2 Population
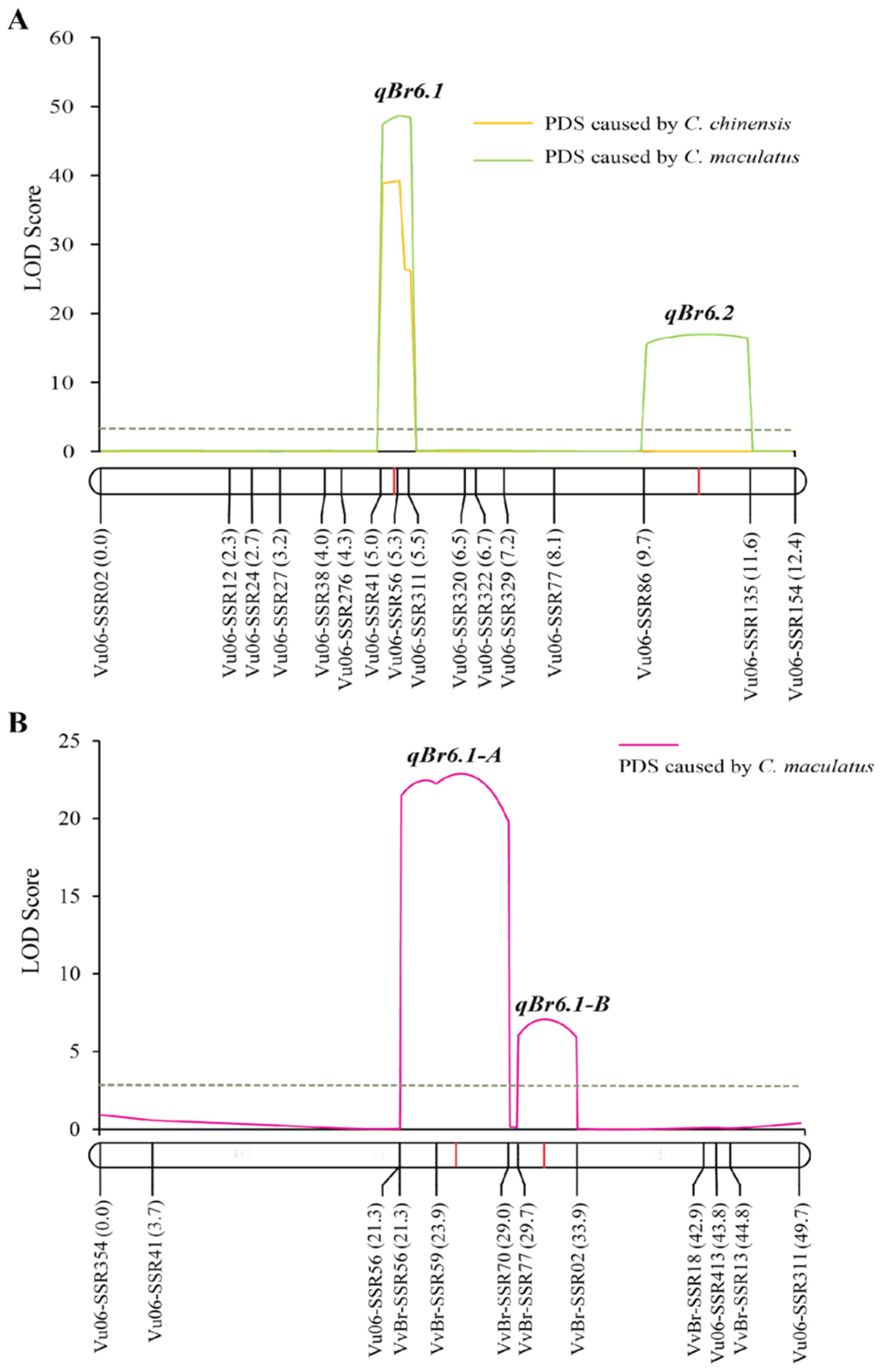
2.2. Fine Mapping qBr6.1 Using the F2 Population
2.3. Variation in Resistance to C. maculatus in the Parents and F2:3 Population
2.4. Narrowing Down the qBr6.1 Region in the F2:3 Population
2.5. Identification of the Candidate Gene for qBr6.1-A and Sequence Variation in the Candidate Gene
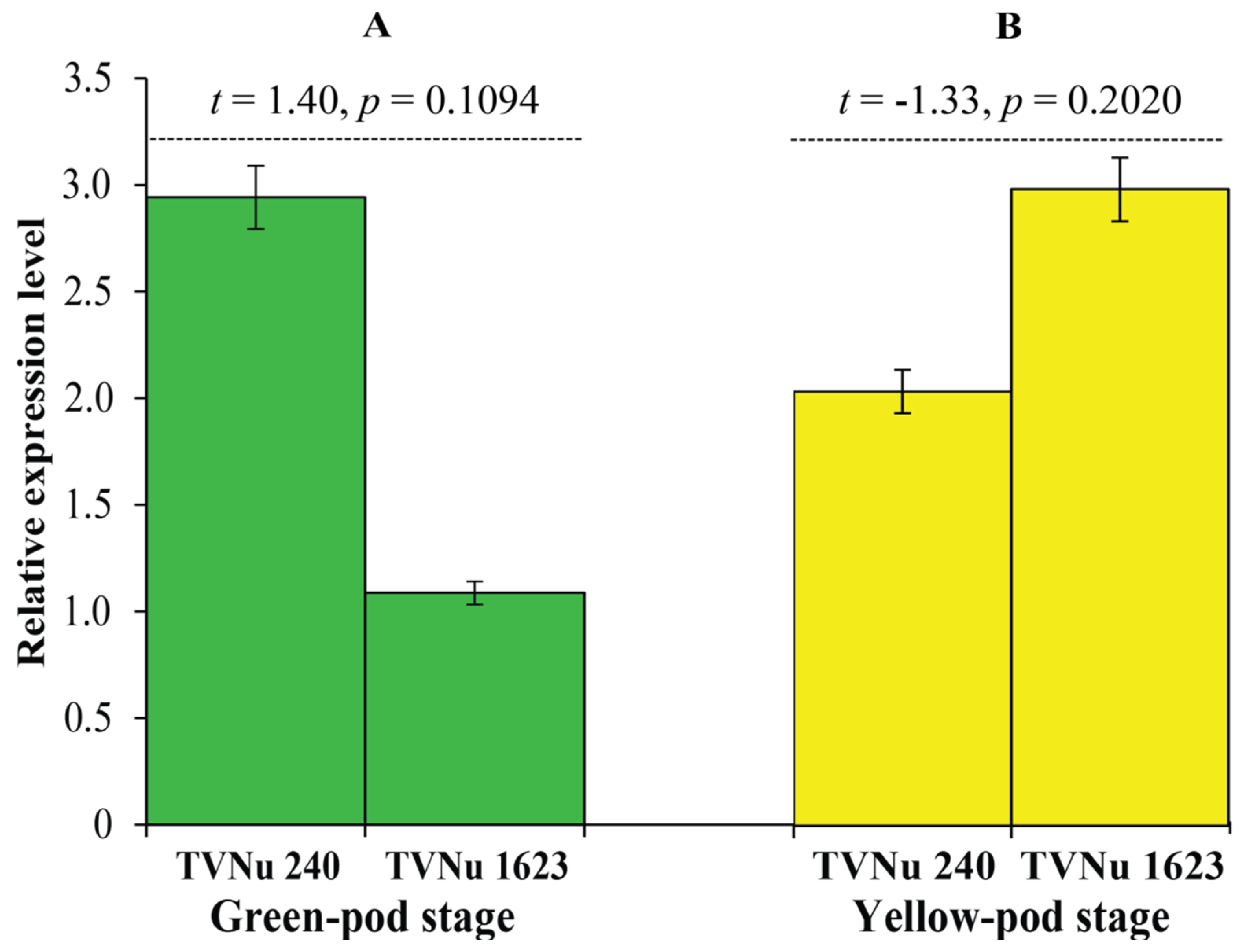
2.6. Expression Analysis of the Gene VvTaXI
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Evaluation of Seed Resistance to Bruchids
4.3. DNA Marker Development and Analysis, and QTL Analysis in the F2 Population
4.4. DNA Marker Development and Analysis, and QTL Analysis in the F2:3 Population
4.5. Identification and Sequencing of a Candidate Gene Controlling Bruchid Resistance
4.6. Gene Expression Analysis of the Candidate Gene
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duke, J.A. Vigna vexillata (L.) Rich. In Handbook of Legumes of World Economic Importance; Plenum Press: New York, NY, USA, 1989; pp. 306–307. [Google Scholar]
- Lawn, R.J.; Williams, R.W.; Imrie, B.C. “Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean”, in Mungbean. In Proceedings of the Second International Symposium, Bangkok, Thailand, 16–20 November 1987; Shanmugasundaram, S., McLean, B.T., Eds.; Asian Vegetable Research and Development Centre: Tainan, Taiwan, 1988; pp. 136–145. [Google Scholar]
- Sasikumar, B.; Sardana, S. Vigna vexillata (Fabaceae), A pulse cum tuber crop of northeastern hill region of India. Econ. Bot. 1988, 42, 292. [Google Scholar]
- Karuniawan, A.; Iswandi, A.; Kale, P.R.; Heinzemann, J.; Grüneberg, W.J. Vigna vexillata (L.) A. Rich. cultivated as a root crop in Bali and Timor. Genet. Resour. Crop Evol. 2006, 53, 213–217. [Google Scholar] [CrossRef]
- Chandel, K.P.S.; Arora, R.K.; Joshi, B.S. Vigna capensis Walp. (V. vexillata) an edible root legume. Curr. Sci. 1972, 41, 537. [Google Scholar]
- Bhattacharyya, P.K.; Ghosh, A.K.; Sanyal, B.; Ray, G.D. Grow Vigna vexillata for protein-rich tuber-cum-pulse crop in North-eastern hill region. Seeds Farms 1984, 10, 33–36. [Google Scholar]
- Dachapak, S.; Somta, P.; Poonchaivilaisak, S.; Yimram, T.; Srinives, P. Genetic diversity and structure of the zombi pea (Vigna vexillata (L.) A. Rich) gene pool based on SSR marker analysis. Genetica 2017, 145, 189–200. [Google Scholar] [CrossRef]
- Southgate, B.J. Biology of the bruchidae. Ann. Rev. Entomol. 1979, 24, 449–473. [Google Scholar] [CrossRef]
- Srinives, S.; Somta, P.; Somta, C. Genetics and breeding of resistance to bruchids (Callosobruchus spp.) in Vigna crops: A review. NU Sci. J. 2007, 4, 1–17. [Google Scholar]
- Deshpande, V.K.; Makanur, B.; Deshpande, S.K.; Adiger, S.; Salimath, P.M. Quantitative and qualitative losses caused by Callosobruchus maculatus in cowpea during seed storage. Plant Arch. 2011, 11, 723–731. [Google Scholar]
- Yadav, P. Susceptibility of four Indian grain legumes to three species of stored pest, bruchid (Callosobruchus) and effect of temperature on bruchids. Int. J. Entomol. Res. 2018, 3, 5–10. [Google Scholar]
- Mishra, S.K.; Macedo, M.L.R.; Panda, S.K.; Panigrahi, J. Bruchid pest management in pulses: Past practices, present status and use of modern breeding tools for development of resistant varieties. Ann. Appl. Biol. 2018, 172, 4–19. [Google Scholar] [CrossRef]
- Madhurya, L.; Nazeer, M.; Urvashi1, S.; Ezhil, V.S. Contamination risk of aluminium phosphide residues on the packed wheat grain sacs. J. Entomol. Res. 2023, 47, 458–460. [Google Scholar] [CrossRef]
- Birch, A.N.E.; Fellows, L.E.; Evans, S.V.; Doherty, K. Para-aminophenylalanine in Vigna: Possible taxonomic and ecological significance as a seed defence against bruchids. Phytochemistry 1986, 25, 2745–2749. [Google Scholar] [CrossRef]
- Bressan, R.A. Contributions of PAPA to V. vexillata resistances: Another opportunity for biotechnology? In Joint Cowpeas Biotechnology Workshop; Purdue University: West Lafayette, IN, USA, 1990; pp. 16–20. [Google Scholar]
- Chotechung, S.; Somta, P.; Chen, J.; Yimram, T.; Chen, X.; Srinives, P.A. Gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016, 129, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Rathnayaka, G.S.; Kaewwongwal, A.; Laosatit, K.; Yimram, T.; Yun, L.; Xin, C.; Nakazono, M.; Somta, P. Tandemly duplicated genes encoding polygalacturonase inhibitors are associated with bruchid (Callosobruchus chinensis) resistance in moth bean (Vigna aconitifolia). Plant Sci. 2022, 323, 111402. [Google Scholar] [CrossRef]
- Amkul, K.; Wang, L.; Somta, P.; Wang, S.; Cheng, X. Construction of a high density linkage map and genome dissection of bruchid resistance in zombi pea (Vigna vexillata (L.) A. Rich). Sci. Rep. 2019, 9, 11719. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, V.; Terzano, R.; Cicco, N.; Cardinali, A.; Venere, D.D.; Linsalata, V. Seed coat tannins and bruchid resistance in stored cowpea seeds. J. Sci. Food Agric. 2005, 85, 839–846. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.-C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef]
- Naito, K.; Wakatake, T.; Shibata, T.F.; Iseki, K.; Shigenobu, S.; Takahashi, Y.; Tanaka, E.O.; Muto, C.; Teruya, K.; Shiroma, A.; et al. Genome sequence of 12 Vigna species as a knowledge base of stress tolerance and resistance. bioRxiv 2020. [Google Scholar] [CrossRef]
- Takahashi, Y.; Somta, P.; Muto, C.; Iseki, K.; Naito, K.; Pandiyan, M.; Senthilkumar, A.N.; Tomooka, N. Novel genetic resources in the genus Vigna unveiled from gene bank accessions. PLoS ONE 2016, 11, e0147568. [Google Scholar] [CrossRef]
- Maréchal, R.; Mascherpa, J.M.; Stainer, F. Etude taxonomique d’un groupe complexe d’speces des genres Phaseolus et Vigna (Papilionaceae) sur la base de donnees morphologiques et polliniques, traitees par l’analyse informatique. Boissiera 1978, 28, 1–273. [Google Scholar]
- Janzen, D.H. Seed-eaters versus seed size, number, toxicity and dispersal. Evolution 1969, 23, 1–27. [Google Scholar] [PubMed]
- Busch, A.; Kunert, G.; Heckel, D.G.; Pauchet, Y. Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles. PLoS ONE 2017, 12, e0184305. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, R.; Kunert, G.; Vogel, H.; Pauchet, Y. Pectin Digestion in Herbivorous Beetles: Impact of Pseudoenzymes Exceeds That of Their Active Counterparts. Front. Physiol. 2019, 10, 685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yan, Q.; Yuan, X.; Lin, Y.; Chen, J.; Wu, R.; Xue, C.; Zhu, Y.; Chen, X. Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J. Plant Physiol. 2021, 258–259, 153376. [Google Scholar] [CrossRef]
- Ishimoto, M.; Kitamura, K. Growth inhibitory effects of an a-amylase inhibitor from the kidney bean, Phaseolus vulgaris (L.) on three species of bruchids (Coleoptera: Bruchidae). Appl. Entomol. Zool. 1989, 24, 281–286. [Google Scholar] [CrossRef]
- Ebringerová, A.; Heinze, T. Xylan and Xylan Derivatives-Biopolymers with Valuable Properties, 1. Naturally Occurring Xylans Structures, Isolation Procedures and Properties. Macromol. Rapid Commun. 2000, 21, 542–556. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Verma, P.A. Detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Tundo, S.; Mandalà, G.; Sella, L.; Favaron, F.; Bedre, R.; Kalunke, R.M. Xylanase inhibitors: Defense players in plant immunity with implications in agro-industrial processing. Int. J. Mol. Sci. 2022, 23, 14994. [Google Scholar] [CrossRef]
- Kouadio, E.J.P.; Konan, K.H.; Djè, K.M.; Dué, E.A.; Kouamé, L.P. Insect digestive glycosidases: Strategies of purification, biochemical properties and potential applications, a review. Int. J. Entomol. Res. 2016, 4, 67–86. [Google Scholar]
- Yan, J.; Chen, J.; Lin, Y.; Yuan, X.; Somta, P.; Zhang, Y.; Chen, X. Mapping of quantitative trait locus reveals PsXI gene encoding xylanase inhibitor as the candidate gene for bruchid (Callosobruchus spp.) resistance in pea (Pisum sativum L.). Front. Plant Sci. 2023, 14, 1057577. [Google Scholar] [CrossRef]
- Fierens, K.; Brijs, K.; Courtin, C.M.; Gebruers, K.; Goesaert, H.; Raedschelders, G.; Robben, J.; Campenhout, S.V.; Volckaert, G.; Delcour, J.A. Molecular identification of wheat endoxylanase inhibitor TAXI-I, member of a new class of plant proteins. FEBS Lett. 2003, 540, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Sansen, S.; De Ranter, C.J.; Gebruers, K.; Brijs, K.; Courtin, C.M.; Delcour, J.A.; Rabijns, A. Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 2004, 279, 36022–36028. [Google Scholar] [CrossRef]
- Pollet, A.; Sansen, S.; Raedschelders, G.; Gebruers, K.; Rabijns, A.; Delcour, J.A.; Courtin, C.M. Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA. FEBS J. 2009, 276, 3916–3927. [Google Scholar] [CrossRef] [PubMed]
- Moscetti, I.; Faoro, F.; Moro, S.; Sabbadin, D.; Sella, L.; Favaron, F.; D’Ovidio, R. The xylanase inhibitor TAXI-III counteracts the necrotic activity of a Fusarium graminearum xylanase in vitro and durum wheat transgenic plants. Mol. Plant Pathol. 2015, 16, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Ye, G.N.; Weeden, N.F.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant. Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3–new capabilities and interfaces. Nucleic. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Laosatit, K.; Amkul, K.; Yimram, T.; Chen, J.; Lin, Y.; Yuan, X.; Wang, L.; Chen, X.; Somta, P. A Class II KNOX Gene, KNAT7-1, Regulates Physical Seed Dormancy in Mungbean [Vigna radiata (L.) Wilczek]. Front. Plant Sci. 2022, 13, 852373. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Van Os, H.; Stam, P.; Visser, R.G.; Van Eck, H.J. RECORD: A novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 2005, 112, 30–40. [Google Scholar] [CrossRef]
- Li, H.; Ye, G.; Wang, J. A modified algorithm for the improvement of composite interval mapping. Genetics 2007, 175, 361–374. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Laksana, C.; Chanprame, S. A simple and rapid method for RNA extraction from young and mature leaves of oil palm (Elaeis guineensis Jacq.). J. ISSAAS 2015, 21, 96–106. [Google Scholar]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆DCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Bruchid Species | QTL Name | Position on Linkage Group 6 (cM) | Marker Interval | LOD Score | PVE (%) | Additive Effect | Dominant Effect |
|---|---|---|---|---|---|---|---|
| C. chinensis | qBr6.1 | 5.3 | Vu06-SSR41–Vu06-SSR56 | 39.20 | 34.78 | −14.07 | −10.43 |
| C. maculatus | qBr6.1 | 5.3 | Vu06-SSR41–Vu06-SSR56 | 48.66 | 34.94 | −29.50 | −1.97 |
| qBr6.2 | 11.2 | Vu06-SSR86–Vu06-SSR135 | 16.93 | 10.42 | −0.25 | −22.86 |
| QTL Name | Position on Linkage Group 6 (cM) | Marker Interval | LOD Score | PVE (%) | Additive Effect | Dominant Effect |
|---|---|---|---|---|---|---|
| qBr6.1-A | 25.6 | VvBr-SSR59–VvBr-SSR70 | 22.89 | 37.46 | −26.59 | −4.38 |
| qBr6.1-B | 31.6 | VvBr-SSR77–VvBr-SSR02 | 7.07 | 10.63 | −0.95 | −26.11 |
| QTL | Gene | Scaffold | Location | Description |
|---|---|---|---|---|
| qBr6.1-A | Vigve.0047s019200.01 | 0047 | 3196221..3196999 (+strand) | Uncharacterized protein |
| Vigve.0047s019300.01 | 0047 | 3199728..3201904 (−strand) | TAXI protein | |
| Vigve.0047s019400.01 | 0047 | 3208183..3217005 (+strand) | Hypothetical protein | |
| Vigve.0047s019500.01 | 0047 | 3222513..3224711 (+strand) | Uncharacterized protein | |
| qBr6.1-B | Vigve.0047s019600.01 | 0047 | 3254888..3256167 (−strand) | Isopenicillin N synthase-like |
| Vigve.0047s019700.01 | 0047 | 3275112..3275345 (−strand) | Hypothetical protein | |
| Vigve.0047s019800.01 | 0047 | 3280299..3281578 (−strand) | Isopenicillin N synthase-like | |
| Vigve.0047s019900.01 | 0047 | 3328148..3348849 (+strand) | Dentin matrix acidic phosphoprotein 1-like | |
| Vigve.0047s020000.01 | 0047 | 3357162..3363147 (−strand) | N-acylphosphatidylethanolamine synthase | |
| Vigve.0047s020100.01 | 0047 | 3372295..3376044 (+strand) | Hypothetical protein | |
| Vigve.0047s020200.01 | 0047 | 3376384..3376962 (−strand) | Protein PXR1 | |
| Vigve.0047s020300.01 | 0047 | 3395240..3404186 (+strand) | Chloroplastic protein TIC 40 | |
| Vigve.0047s020400.01 | 0047 | 3404965..3407557 (+strand) | Uncharacterized protein | |
| Vigve.0047s020500.01 | 0047 | 3407911..3411731 (−strand) | WAT1-related protein At3g02690 | |
| Vigve.0047s020600.01 | 0047 | 3414400..3416005 (+strand) | Late embryogenesis abundant protein | |
| Vigve.0047s020700.01 | 0047 | 3417441..3419499 (+strand) | Protein ROOT PRIMORDIUM DEFECTIVE 1 | |
| Vigve.0047s020800.01 | 0047 | 3421598..3422730 (−strand) | Uncharacterized protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amkul, K.; Laosatit, K.; Lin, Y.; Yuan, X.; Chen, X.; Somta, P. A Gene Encoding Xylanase Inhibitor Is a Candidate Gene for Bruchid (Callosobruchus spp.) Resistance in Zombi Pea (Vigna vexillata (L.) A. Rich). Plants 2023, 12, 3602. https://doi.org/10.3390/plants12203602
Amkul K, Laosatit K, Lin Y, Yuan X, Chen X, Somta P. A Gene Encoding Xylanase Inhibitor Is a Candidate Gene for Bruchid (Callosobruchus spp.) Resistance in Zombi Pea (Vigna vexillata (L.) A. Rich). Plants. 2023; 12(20):3602. https://doi.org/10.3390/plants12203602
Chicago/Turabian StyleAmkul, Kitiya, Kularb Laosatit, Yun Lin, Xingxing Yuan, Xin Chen, and Prakit Somta. 2023. "A Gene Encoding Xylanase Inhibitor Is a Candidate Gene for Bruchid (Callosobruchus spp.) Resistance in Zombi Pea (Vigna vexillata (L.) A. Rich)" Plants 12, no. 20: 3602. https://doi.org/10.3390/plants12203602





