An Integrated Analysis of microRNAs and the Transcriptome Reveals the Molecular Mechanisms Underlying the Regulation of Leaf Development in Xinyang Maojian Green Tea (Camellia sinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction
2.3. sRNA and RNA Sequencing (RNA-Seq) Library Construction and Sequencing
2.4. Identification of Conserved and Novel miRNAs
2.5. Annotation and Cluster Analysis of miRNAs
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
2.7. qRT-PCR Validation of miRNAs
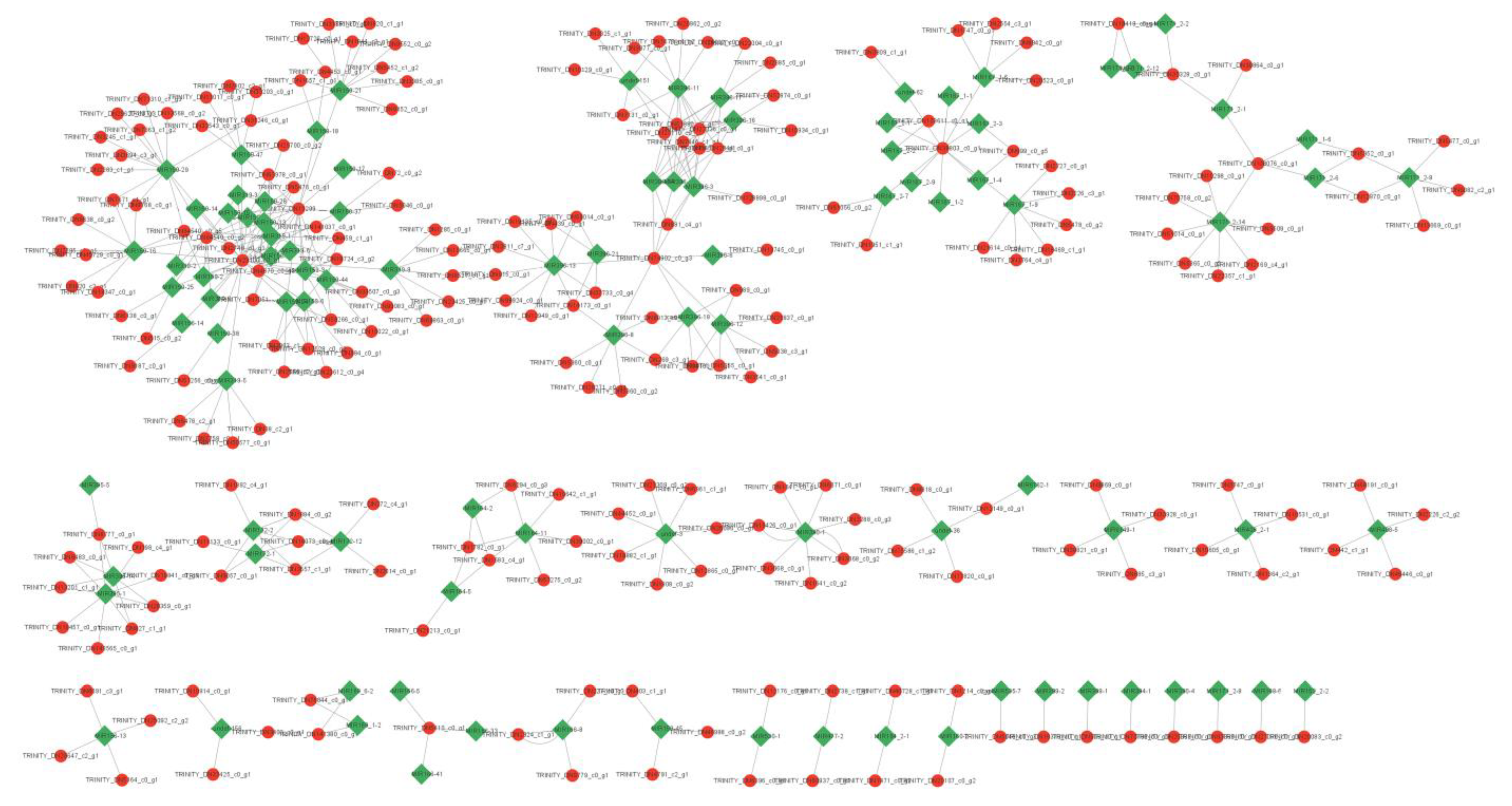
2.8. Target Prediction and Construction of a Regulatory and Interaction Network
2.9. Statistical Analysis
3. Results
3.1. High-Throughput Sequencing of sRNAs in XYMJ Tea Plants
3.2. Conserved miRNAs in XYMJ Tea Plants
3.3. Characterization of Conserved miRNAs
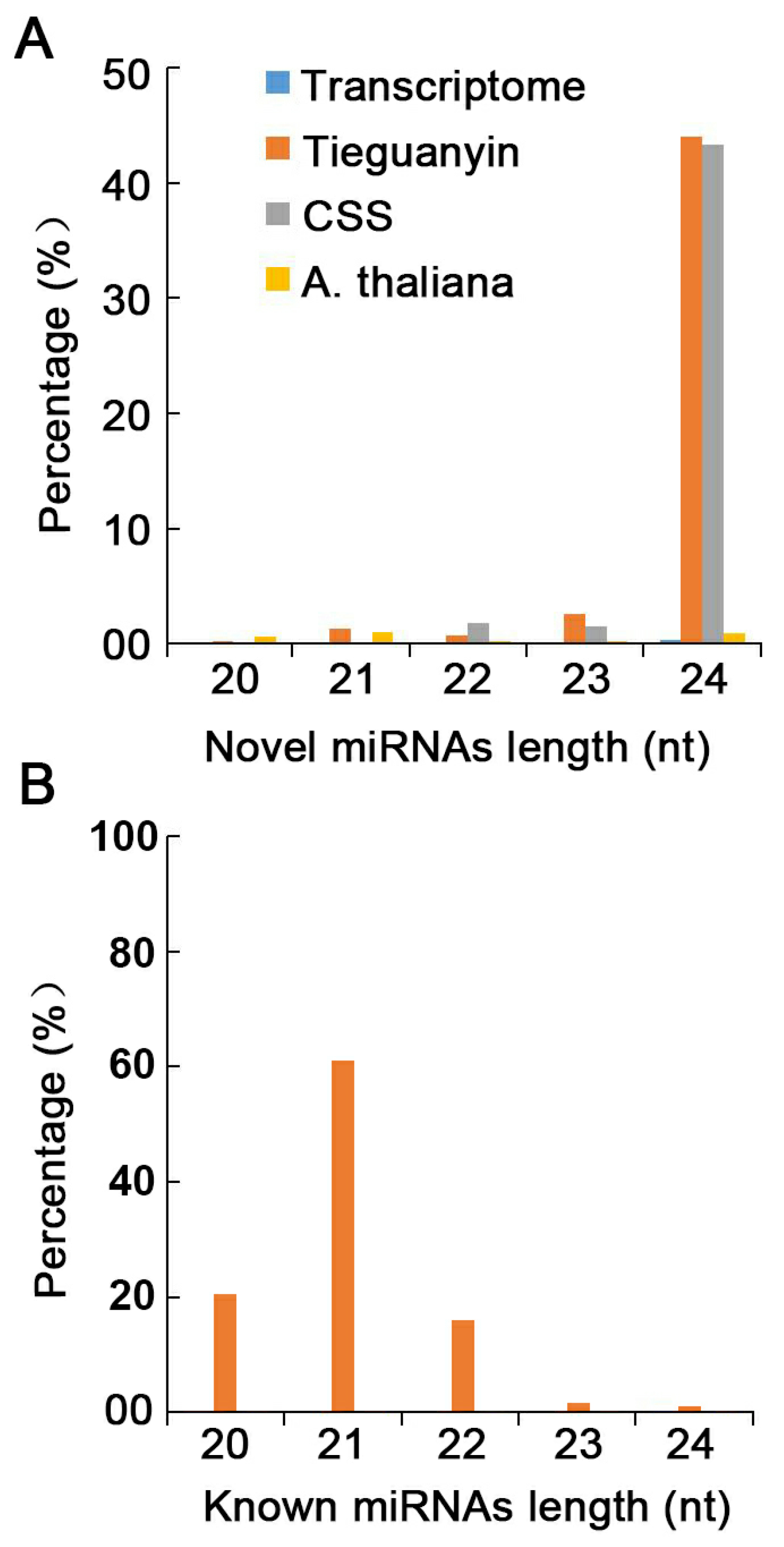
3.4. Identification of Novel miRNAs in XYMJ Tea Plants
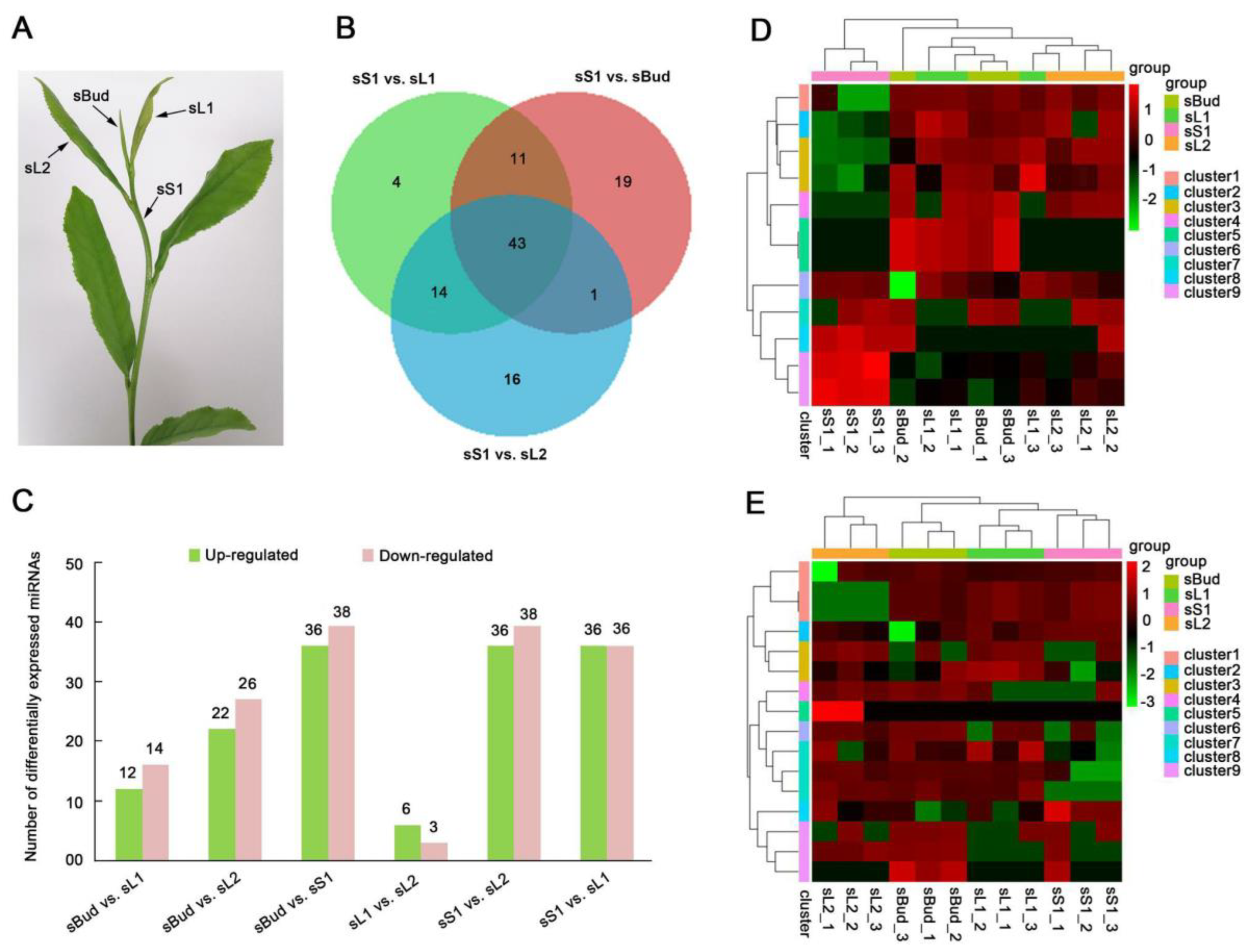
3.5. Tissue-Specific Expression of miRNAs
3.6. Analysis of DEMs Involved in Leaf Development
3.7. DEG Analysis
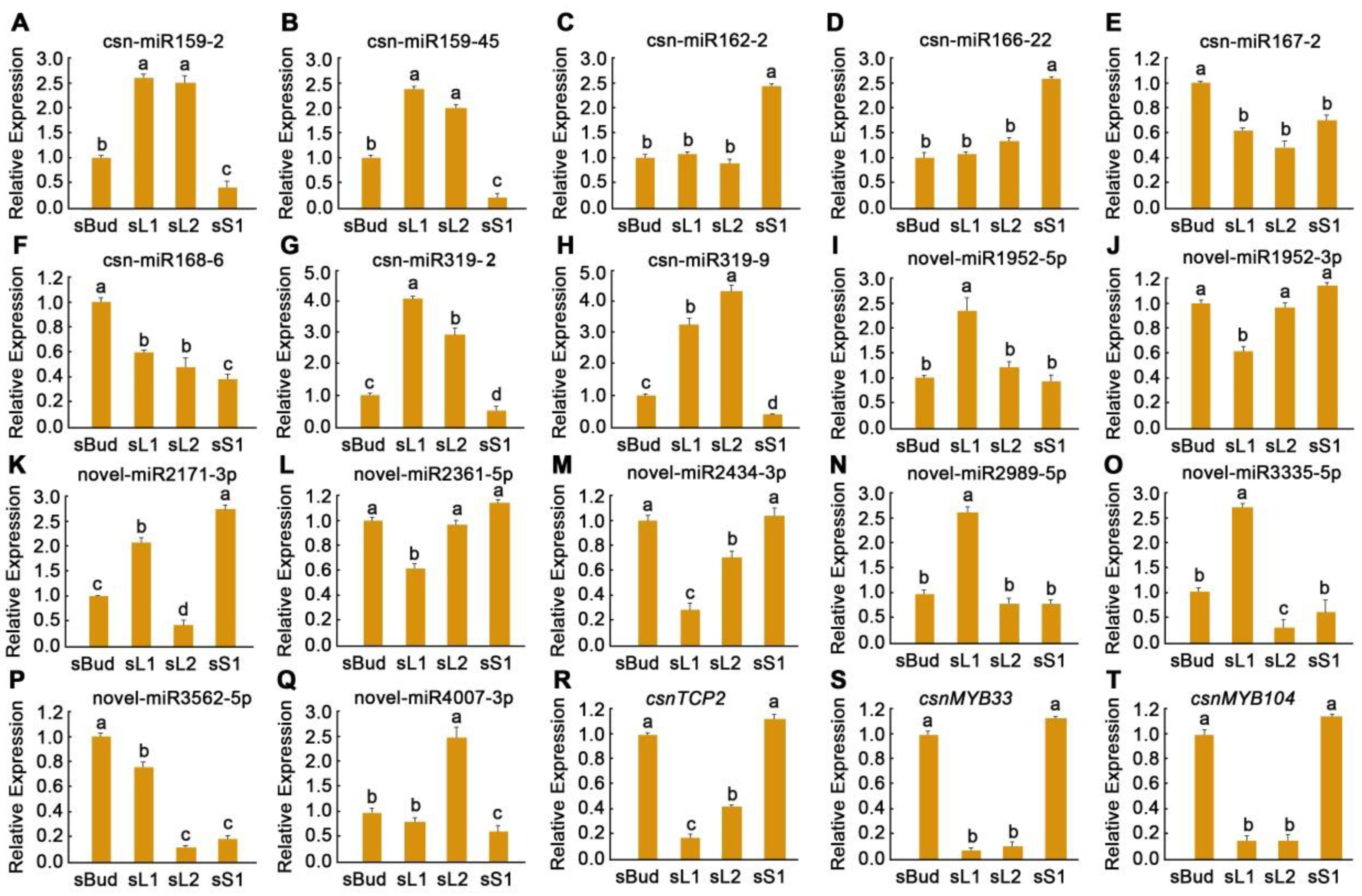
3.8. Validation of Gene Expression by qRT-PCR
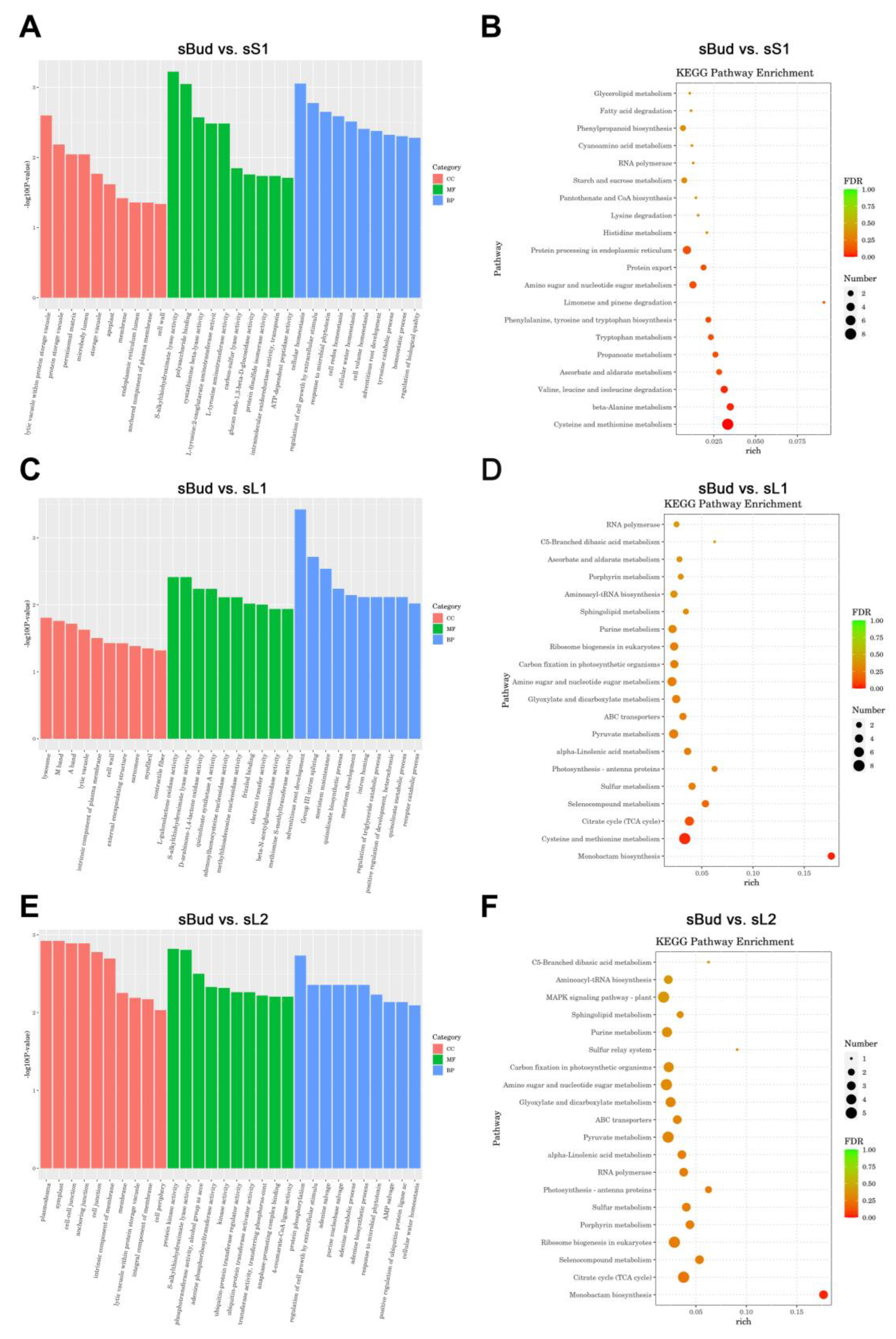
3.9. GO and KEGG Analyses
3.10. Key DEMs Involved in Leaf Growth and Development
3.11. Expression Analysis of miRNAs and Their Target mRNAs
4. Discussion
4.1. miRNAs Involved in the Development of the Leaves of XYMJ Tea Plants Were Expressed in Different Tissues and Developmental Stages
4.2. Differentially Expressed Transcription Factors (TFs) Involved in the Development of the Leaves of XYMJ Tea Plants
4.3. Network of miRNAs and mRNAs Involved in Regulating Leaf Development in XYMJ Tea Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA. 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.Z.; Wei, C.L.; Wan, X.C. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2000, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Ming, T.; Bartholomew, B. Theaceae. In Flora of China; Wu, Z., Raven, P., Hong, D., Eds.; Science Press and Missouri Botanical Garden: Beijing, China; St. Louis, MI, USA, 2007; pp. 367–412. [Google Scholar]
- Kaundun, S.S.; Matsumoto, S. Development of CAPS markers based on three key genes of the phenylpropanoid pathway in tea, Camellia sinensis (L.) O. Kuntze, and differentiation between assamica and sinensis varieties. Theor Appl. Genet. 2003, 106, 375–383. [Google Scholar] [CrossRef]
- Mukhopadhyay, M.; Mondal, T.K.; Chand, P.K. Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): A review. Plant Cell Rep. 2016, 35, 255–287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, C.; Wang, Y.; Shen, J.; Ding, Z. Conserved MicroRNA act boldly during sprout development and quality formation in Pingyang Tezaocha (Camellia sinensis). Front. Genet. 2019, 10, 237. [Google Scholar] [CrossRef]
- Zhou, C.; Tian, C.; Zhu, C.; Lai, Z.; Lin, Y.; Guo, Y. Hidden players in the regulation of secondary metabolism in tea plant: Focus on non-coding RNAs. Beverage Plant Res. 2022, 2, 19. [Google Scholar] [CrossRef]
- Lei, X.; Wang, Y.; Zhou, Y.; Chen, Y.; Chen, H.; Zou, Z.; Zhou, L.; Ma, Y.; Chen, F.; Wang, W. TeaPGDB: Tea Plant Genome Database. Beverage Plant Res. 2021, 1, 5. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.Y.; Chu, C.C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; You, C.; Chen, X. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 2017, 35, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. Control of cell proliferation by microRNAs in plants. Curr. Opin. Plant Biol. 2016, 34, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Fouracre, J.P.; Poethig, R.S. The role of small RNAs in vegetative shoot development. Curr. Opin. Plant Biol. 2016, 29, 64–72. [Google Scholar] [CrossRef]
- Zheng, C.; Ma, J.Q.; Ma, C.L.; Yao, M.Z.; Chen, J.D.; Chen, L. Identifying conserved functional gene modules underlying the dynamic regulation of tea plant development and secondary metabolism. J. Agric. Food Chem. 2020, 68, 11026–11037. [Google Scholar] [CrossRef]
- Li, P.; Xia, E.; Fu, J.; Xu, Y.; Zhao, X.; Tong, W. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 2022, 110, 1144–1165. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and function of miR159 in Plants. Plants 2019, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A regulatory network for miR156-SPL module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Hao, K.; Wang, Y.; Zhu, Z.; Wu, Y.; Chen, R.; Zhang, L. miR160: An Indispensable Regulator in Plant. Front. Plant Sci. 2022, 13, 833322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Pan, X.P.; Wang, Q.L.; Cobb, G.P.; Anderson, T.A. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005, 15, 336–360. [Google Scholar] [CrossRef] [PubMed]
- Prabu, G.R.; Mandal, A.K. Computational identification of miRNAs and their target genes from expressed sequence tags of tea (Camellia sinensis). Genom. Proteom. Bioinform. 2010, 8, 113–121. [Google Scholar] [CrossRef]
- Jeyaraj, A.; Elango, T.; Li, X.; Guo, G. Utilization of microRNAs and their regulatory functions for improving biotic stress tolerance in tea plant [Camellia sinensis (L.) O. Kuntze]. RNA Biol. 2020, 10, 1365–1382. [Google Scholar] [CrossRef]
- Zhao, J.; Li, P.; Xia, T.; Wan, X. Exploring plant metabolic genomics: Chemical diversity, metabolic complexity in the biosynthesis and transport of specialized metabolites with the tea plant as a model. Crit. Rev. Biotechnol. 2020, 40, 667–688. [Google Scholar] [CrossRef]
- Mohanpuria, P.; Yadav, S.K. Characterization of novel small RNAs from tea (Camellia sinensis L.). Mol. Biol. Rep. 2012, 39, 3977–3986. [Google Scholar] [CrossRef]
- Jeyaraj, A.; Zhang, X.; Hou, Y.; Shangguan, M.; Gajjeraman, P.; Li, Y.; Wei, C. Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant Biol. 2017, 17, 212. [Google Scholar] [CrossRef]
- Liu, S.; Mi, X.; Zhang, R.; An, Y.; Zhou, Q.; Yang, T.; Xia, X.; Guo, R.; Wang, X.; Wei, C. Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis). Planta 2019, 250, 1111–1129. [Google Scholar] [CrossRef]
- Liu, S.; An, Y.; Tong, W.; Qin, X.; Samarina, L.; Guo, R.; Xia, X.; Wei, C. Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis) and development of InDel markers for genetic research. BMC Genom. 2019, 20, 935. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Guo, L.; Shen, J.; Hu, H.; Zhou, R. Transcriptome analysis of Crimson seedless grapevine (Vitis vinifera L.) infected by grapevine berry inner necrosis virus. Curr. Res. Virol. Sci. 2022, 3, 100024. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Guo, Y.; Wang, P.; Wang, Y.; Ni, D. Transcriptional profiling of catechins biosynthesis genes during tea plant leaf development. Planta 2017, 246, 1139–1152. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Ma, Y.K.; Chen, T.; Wang, M.; Wang, X.J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, 22–28. [Google Scholar] [CrossRef]
- Song, H.; Zhang, X.; Shi, C.; Wang, S.S.; Wu, A.; Wei, C. Selection and verification of candidate reference genes for mature microRNA expression by quantitative RT-PCR in the tea plant (Camellia sinensis). Genes 2016, 7, 25. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Zhu, C.; Chang, X.; Yue, C.; Wang, Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Bio. 2017, 17, 211. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 35, 21–44. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs in development - insights from plants. Curr. Opin. Genet. Dev. 2012, 22, 361–367. [Google Scholar] [CrossRef]
- Qi, J.; Wu, B.; Feng, S.; Lü, S.; Guan, C.; Zhang, X.; Qiu, D.; Hu, Y.; Zhou, Y.; Li, C.; et al. Mechanical regulation of organ asymmetry in leaves. Nat. Plants 2017, 3, 724–733. [Google Scholar] [CrossRef]
- Kidner, C.A. The many roles of small RNAs in leaf development. J. Genet. Genomics 2010, 37, 13–21. [Google Scholar] [CrossRef]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Pang, M.; Agarwal, V.; Chen, Z.J. Interspecies regulation of microRNAs and their targets. Biochim. Biophys Acta 2008, 1779, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, X.; Yan, X.; Guo, L.; Mi, X.; Xu, Q.; Zhu, J.; Wu, A.; Liu, L.; Wei, C. Revealing of microRNA involved regulatory gene networks on terpenoid biosynthesis in Camellia sinensis in different growing time points. J. Agric. Food Chem. 2018, 66, 12604–12616. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, Q.Q.; Yan, M.L.; Wang, M.L.; Wang, P.; Zhao, H.; Wang, Y.; Ni, D.J.; Guo, F. Relationship between secondary metabolism and miRNA for important flavor compounds in different tissues of tea plant (Camellia sinensis) as revealed by genome-wide miRNA analysis. J. Agric. Food Chemisrty 2021, 69, 2001–2012. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- He, Z.; Liu, C.; Zhang, Z.; Wang, R.; Chen, Y. Integration of mRNA and miRNA analysis reveals the differentially regulatory network in two different Camellia oleifera cultivars under drought stress. Front. Plant Sci. 2022, 13, 1001357. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Kryovrysanaki, N.; James, A.; Bardani, E.; Kalantidis, K. RNA silencing pathways in plant development and defense. Int. J. Dev. Biol. 2022, 66, 163. [Google Scholar] [CrossRef]
- Allen, R.S.; Li, J.; Stahle, M.I.; Dubroué, A.; Gubler, F.; Millar, A.A. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 2007, 104, 16371–16376. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef] [PubMed]

| Sample | rRNA | snoRNA | snRNA | tRNA |
|---|---|---|---|---|
| sBud-1 | 290,670 | 15,355 | 10,113 | 44,202 |
| sBud-2 | 266,525 | 13,058 | 8667 | 38,303 |
| sBud-3 | 223,086 | 13,483 | 9053 | 34,475 |
| sL1-1 | 250,434 | 10,991 | 7016 | 33,634 |
| sL1-2 | 300,386 | 10,672 | 7370 | 38,165 |
| sL1-3 | 246,187 | 10,370 | 6873 | 31,690 |
| sS1-1 | 227,361 | 10,842 | 7439 | 25,658 |
| sS1-2 | 243,849 | 11,896 | 8133 | 28,680 |
| sS1-3 | 252,037 | 12,271 | 8102 | 28,274 |
| sL2-1 | 350,595 | 11,119 | 7909 | 38,158 |
| sL2-2 | 298,649 | 11,791 | 8068 | 33,514 |
| sL2-3 | 299,478 | 11,342 | 7768 | 32,341 |
| Total | 3,249,257 | 143,190 | 96,511 | 407,094 |
| Comparison of Databases | Transcriptome | Tieguanyin | CSS | Arabidopsis |
|---|---|---|---|---|
| Known miRNA NO./novel miRNA NO. | 381/4 | 381/506 | 381/485 | 381/32 |
| Predicted target genes No. of miRNAs/novel miRNA NO. | 107/0 | 103/2 | 104/7 | 99/3 |
| No. of target genes in known miRNAs/novel miRNA | 703/0 | 510/12 | 537/45 | 348/18 |
| Predicted target genes locus No. of known miRNAs/novel miRNA | 1116/0 | 861/12 | 864/56 | 1224/32 |
| miRNA | Expression Pattern | Target Genes |
|---|---|---|
| csn-miR156-14 | Downregulated (sBud/sS1, sBud/sL1, sBud/sL2) | AT5G50590, AT5G50690 |
| csn-miR156-26 | Upregulated (sBud/sS1, sBud/sL2) | AT5G50960, AT3G14270, AT2G45820 |
| csn-miR159-2 | Upregulated (sBud/sS1), downregulated (sBud/sL1, sBud/sL2) | AT5G67090, AT3G06450, AT2G26950 |
| csn-miR159-45 | Upregulated (sBud/sS1), downregulated (sBud/sL1, sBud/sL2) | AT3G57060, AT2G03250, AT4G18390, AT2G13610, AT3G19930, AT2G43430 |
| csn-miR160-2 | Downregulated (sBud/sS1, sBud/sL1, sBud/sL2), | AT1G77850, AT2G28350, AT4G30080 |
| csn-miR162-2 | downregulated (sBud/sS1) | AT3G01330, AT2G23180 |
| csn-miR166-22 | Downregulated (sBud/sS1, sBud/sL2) | AT1G07810, AT1G30490, AT1G52150 |
| csn-miR167-2 | Upregulated (sBud/sS1, sBud/sL1, sBud/sL2) | AT5G41300, AT2G48110, AT3G57800 |
| csn-miR168-6 | Upregulated (sBud/sS1) | AT5G07140, AT5G17780, AT3G07195 |
| csn-miR319-2 | Upregulated (sBud/sS1), downregulated (sBud/sL1, sBud/sL2) | AT3G57090, AT3G61850, AT2G07689 |
| csn-miR319-9 | Upregulated (sBud/sS1), downregulated (sBud/sL1, sBud/sL2) | AT5G14820, AT5G46530, AT5G56970, AT3G30739, AT3G32980, AT3G62470 |
| novel-miR3335-5p | Downregulated (sBud/sS1) | |
| novel-miR3562-5p | Upregulated (sBud/sS1, sBud/sL1, sBud/sL2) | |
| novel-miR4007-3p | Downregulated (sBud/sL2) | |
| novel-miR4150-5p | Downregulated (sBud/sL1) | CAS037071, CAS096907 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhou, R.; Zhao, S.; Niu, S. An Integrated Analysis of microRNAs and the Transcriptome Reveals the Molecular Mechanisms Underlying the Regulation of Leaf Development in Xinyang Maojian Green Tea (Camellia sinensis). Plants 2023, 12, 3665. https://doi.org/10.3390/plants12213665
Wang X, Zhou R, Zhao S, Niu S. An Integrated Analysis of microRNAs and the Transcriptome Reveals the Molecular Mechanisms Underlying the Regulation of Leaf Development in Xinyang Maojian Green Tea (Camellia sinensis). Plants. 2023; 12(21):3665. https://doi.org/10.3390/plants12213665
Chicago/Turabian StyleWang, Xianyou, Ruijin Zhou, Shanshan Zhao, and Shengyang Niu. 2023. "An Integrated Analysis of microRNAs and the Transcriptome Reveals the Molecular Mechanisms Underlying the Regulation of Leaf Development in Xinyang Maojian Green Tea (Camellia sinensis)" Plants 12, no. 21: 3665. https://doi.org/10.3390/plants12213665
APA StyleWang, X., Zhou, R., Zhao, S., & Niu, S. (2023). An Integrated Analysis of microRNAs and the Transcriptome Reveals the Molecular Mechanisms Underlying the Regulation of Leaf Development in Xinyang Maojian Green Tea (Camellia sinensis). Plants, 12(21), 3665. https://doi.org/10.3390/plants12213665





