The Role of Cytokinins during the Development of Strawberry Flowers and Receptacles
Abstract
:1. Introduction
2. Results
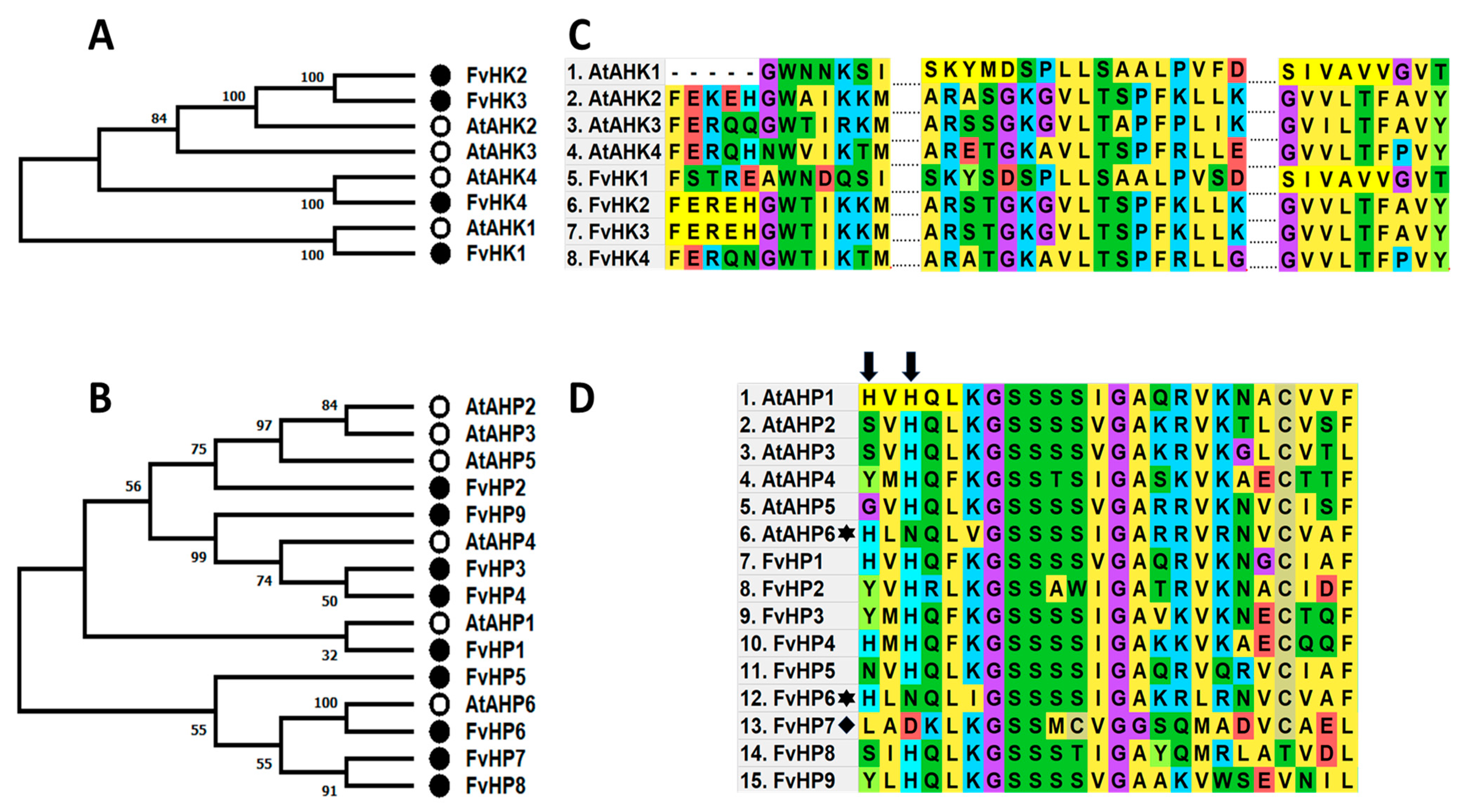
2.1. Cytokinin Genes Are Expressed during Flower and Receptacle Development
2.2. Cytokinin Content Diminishes after Anthesis
2.3. Exogenous Application of Cytokinins in Strawberry Plants Impact Flower Number and Fruit Size and Weight
2.4. Spatio-Temporal Gene Expression Analysis of CKX Genes in Developing Receptacles of F. × ananassa
3. Discussion
3.1. Cytokinin Related Gene Expression and Cytokinin Content Change during Flower and Receptacle Development
3.2. Cytokinin Concentration
3.3. Exogenous Application in the Field Led to Changes in Flower Number and Fruit Size and Weight
3.4. Spatiotemporal Expression Analysis of Cytokinin Pathway Genes in Developing Receptacles of F. × ananassa
4. Materials and Methods
4.1. Biological Material
4.2. Gene Identification and In Silico Analyses
4.3. Cytokinin Determination
4.4. Exogenous Application of Cytokinins in Strawberry Plants
4.4.1. Statistical Analysis
4.4.2. Spatio-Temporal Expression Analysis (In Situ Hybridization)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prámparo, M.B.; Quattrocchio, M.; Gandolfo, M.A.; Del, M.; Zamaloa, C.; Paleontológico, M.; Feruglio, E. Historia Evolutiva de Las Angiospermas (Cretácico-Paleógeno) En Argentina a Través de Los Registros Paleoflorísticos. Asoc. Paleontológica Argentina 2007, 11, 157–172. [Google Scholar]
- Ferrandiz, C. Fruit Development: Turning Sticks into Hearts. Curr. Biol. 2019, 29, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Sotelo-Silveira, M.; Marsch-Martínez, N.; de Folter, S. Unraveling the Signal Scenario of Fruit Set. Planta 2014, 239, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Lenhard, M. Control of Organ Size in Plants. Curr. Biol. 2012, 22, 360–367. [Google Scholar] [CrossRef]
- Ozga, J.A.; Reinecke, D.M. Hormonal Interactions in Fruit Development. J. Plant Growth Regul. 2003, 22, 73–81. [Google Scholar] [CrossRef]
- Skoog, F.; Miller, C.O. Chemical Regulation of Growth and Organ Formation in Plant Tissues Cultured in Vitro. Symp. Soc. Exp. Biol. 1957, 11, 118130. [Google Scholar]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of Plant Growth by Cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis Thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Zuñiga-Mayo, V.M.; Baños-Bayardo, C.R.; Díaz-Ramírez, D.; Marsch-Martínez, N.; De Folter, S. Conserved and Novel Responses to Cytokinin Treatments during Flower and Fruit Development in Brassica napus and Arabidopsis thaliana. Sci. Rep. 2018, 8, 6836. [Google Scholar] [CrossRef]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; De Folter, S. The Role of Cytokinin during Arabidopsis gynoecia and Fruit Morphogenesis and Patterning. Plant J. 2012, 72, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Marsch-Martínez, N.; de Folter, S. Hormonal Control of the Development of the Gynoecium. Curr. Opin. Plant Biol. 2016, 29, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.J.; Larsson, E.; Spíchal, L.; Sundberg, E. Cytokinin-Auxin Crosstalk in the Gynoecial Primordium Ensures Correct Domain Patterning 1[OPEN]. Plant Physiol. 2017, 175, 1144–1157. [Google Scholar] [CrossRef] [PubMed]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL Controls Meristem Function by Direct Regulation of Cytokinin-Inducible Response Regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Chickarmane, V.S.; Gordon, S.P.; Tarr, P.T.; Heisler, M.G.; Meyerowitz, E.M. Cytokinin Signaling as a Positional Cue for Patterning the Apical-Basal Axis of the Growing Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2012, 109, 4002–4007. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI Proteins Activate Cytokinin Biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-Mediated Cytokinin Transcriptional Network Directs Hormone Cross-Regulation and Shoot Development. Nat. Commun. 2018, 9, 1–13. [Google Scholar]
- Kieber, J.J.; Schaller, E.G. Cytokinin Signaling in Plant Development. Development 2018, 145, 1–7. [Google Scholar] [CrossRef]
- Heyl, A.; Werner, T.; Schmülling, T. Cytokinin Signal Perception and Transduction. Curr. Opin. Plant Biol. 2018, 6, 480–488. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, Biosynthesis, and Translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin Glucosyl Transferases, Key Regulators of Cytokinin Homeostasis, Have Potential Value for Wheat Improvement. Plant Biotechnol. J. 2021, 19, 878–896. [Google Scholar] [CrossRef]
- Liu, C.J.; Zhao, Y.; Zhang, K. Cytokinin Transporters: Multisite Players in Cytokinin Homeostasis and Signal Distribution. Front. Plant Sci. 2019, 10, 693. [Google Scholar] [CrossRef] [PubMed]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Bartrina, I. Structure and Function of Cytokinin Oxidase/Dehydrogenase Genes of Maize, Rice, Arabidopsis and Other Species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, I.; Scheirlinck, M.T.; Otto, E.; Bartrina, I.; Schmidt, R.C.; Schmülling, T. Cytokinin Regulates the Activity of the Inflorescence Meristem and Components of Seed Yield in Oilseed Rape. J. Exp. Bot. 2020, 71, 7146–7159. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin Oxidase Regulates Rice Grain Production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of the HvCKX1 Gene Decreases the Cytokinin Oxidase/Dehydrogenase Level in Barley and Leads to Higher Plant Productivity. J. Exp. Bot. 2010, 61, 1839–1851. [Google Scholar] [CrossRef]
- Zhao, J.; Bai, W.; Zeng, Q.; Song, S.; Zhang, M.; Li, X.; Hou, L.; Xiao, Y.; Luo, M.; Li, D.; et al. Moderately Enhancing Cytokinin Level by Down-Regulation of GhCKX Expression in Cotton Concurrently Increases Fiber and Seed Yield. Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef]
- Hancock, J.F.; Sjulin, T.M.; Lobos, G.A. Temperate Fruit Crop Breeding: Germplasm to Genomics. Strawberries. In Temperate Fruit Crop Breeding: Germplasm to Genomics; Hancok, J.F., Ed.; Springer, Kluwer Academic Publishers: Dordrecht, The Netherlands, 2008; pp. 393–437. [Google Scholar]
- Hawkins, C.; Caruana, J.; Li, J.; Zawora, C.; Darwish, O.; Wu, J.; Alkharouf, N.; Liu, Z. An EFP Browser for Visualizing Strawberry Fruit and Flower Transcriptomes. Hortic. Res. 2017, 4, 1–8. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Carvalho, S.D.; O’Grady, K.; Folta, K.M. Agroinfiltration of Strawberry Fruit—A Powerful Transient Expression System for Gene Validation. Curr. Plant Biol. 2016, 6, 19–37. [Google Scholar] [CrossRef]
- Van Bel, M.; Silvestri, F.; Weitz, E.M.; Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K. PLAZA 5.0: Extending the Scope and Power of Comparative and Functional Genomics in Plants. Nucleic Acids Res. 2022, 50, 1468–1474. Available online: https://bioinformatics.psb.ugent.be/plaza/ (accessed on 20 March 2021). [CrossRef]
- Mi, X.; Wang, X.; Wu, H.; Gan, L.; Ding, J.; Li, Y. Characterization and Expression Analysis of Cytokinin Biosynthesis Genes in Fragaria Vesca. Plant Growth Regul. 2017, 82, 139–149. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, Y.; Mi, X.; Gan, L.; Gu, T.; Ding, J.; Li, Y. Identification and Expression Analysis of Cytokinin Response Regulators in Fragaria Vesca. Acta Physiol. Plant 2016, 38, 198. [Google Scholar] [CrossRef]
- Jiang, Y.; Mi, X.; Lin, Y.; Wu, H.; Gu, T.; Ding, J.; Li, Y. Evolution and Expression Patterns of Cytokinin Oxidase Genes in Fragaria vesca. Sci. Hortic. 2016, 212, 115–125. [Google Scholar] [CrossRef]
- Heyl, A.; Wulfetange, K.; Pils, B.; Nielsen, N.; Romanov, G.A.; Schmülling, T. Evolutionary Proteomics Identifies Amino Acids Essential for Ligand-Binding of the Cytokinin Receptor CHASE Domain. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Yakubov, B.; Satoh, R.; Yamaguchi-Shinozaki, K.; Seki, M.; Hirayama, T.; Shinozaki, K. A Transmembrane Hybrid-Type Histidine Kinase in Arabidopsis Functions as an Osmosensor. Plant Cell 1999, 11, 1743–1754. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of AHK1/ATHK1 and Cytokinin Receptor Histidine Kinases in Response to Abscisic Acid, Drought, and Salt Stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Ariza, M.T.; Soria, C.; Martínez-Ferri, E. Developmental Stages of Cultivated Strawberry Flowers in Relation to Chilling Sensitivity. AoB Plants 2015, 7, plv012. [Google Scholar] [CrossRef]
- Hollender, C.A.; Geretz, A.C.; Slovin, J.P.; Liu, Z. Flower and Early Fruit Development in a Diploid Strawberry, Fragaria vesca. Planta 2012, 235, 1123–1139. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and Evolution of the Octoploid Strawberry Genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Hwang, I.; Chen, H.C.; Sheen, J. Two-Component Signal Transduction Pathways in Arabidopsis. Plant Physiol. 2002, 129, 500–515. [Google Scholar] [CrossRef]
- Jin, S.H.; Ma, X.M.; Kojima, M.; Sakakibara, H.; Wang, Y.W.; Hou, B.K. Overexpression of Glucosyltransferase UGT85A1 Influences Trans-Zeatin Homeostasis and Trans-Zeatin Responses Likely through O-Glucosylation. Planta 2013, 237, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Chiang, Y.H.; Kieber, J.J.; Schaller, G.E. SCFKMD Controls Cytokinin Signaling by Regulating the Degradation of Type-B Response Regulators. Proc. Natl. Acad. Sci. USA 2013, 110, 10028–10033. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Goertzen, L.R. The CRF Domain Defines Cytokinin Response Factor Proteins in Plants. BMC Plant Biol. 2010, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; Jia, S.; Huang, X.; Wang, L.; Fu, W.; Huo, G.; Gan, L.; Ding, J.; Li, Y. Transcriptome and Hormone Analyses Provide Insights into Hormonal Regulation in Strawberry Ripening. Planta 2019, 250, 145–162. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Motyka, V.; Pokorna, E.; Dobrev, P.I.; Lacek, J.; Shao, J.; Lewers, K.S.; Mattoo, A.K. Comprehensive Profiling of Endogenous Phytohormones and Expression Analysis of 1-Aminocyclopropane-1-Carboxylic Acid Synthase Gene Family during Fruit Development and Ripening in Octoploid Strawberry (Fragaria × Ananassa). Plant Physiol. Biochem. 2023, 196, 186–196. [Google Scholar] [CrossRef]
- Yang, L.; Jon, C.S.; Wang, L.; Zou, Y.; Liu, L.; Ri, H.C.; Zhao, J.; Cui, M.; Shang, H.; Li, D. Analysis of Multiple-Phytohormones during Fruit Development in Strawberry by Using Miniaturized Dispersive Solid-Phase Extraction Based on Ionic Liquid-Functionalized Carbon Fibers. J. Food Compos. Anal. 2022, 106, 104262. [Google Scholar] [CrossRef]
- Al-Madhagi, I.; Hasan, S.M.Z.; Ahmad, A.; Abdulbaset, I.; Al-Madhagi, H.; Bin Ahmad, A.; Zain, A.M.; Abdullah Bin Yusoff, W. The Influence of Exogenous Hormone on the Flowering and Fruiting of Strawberry (Fragaria × Ananassa Duch). J. Biol. Agric. Healthc. 2012, 2, 46–52. [Google Scholar]
- Takur, Y.; Chandel, J.S.; Verma, P. Effect of Plant Growth Regulators on Growth, Yield and Fruit Quality of strawberry (Fragaria × Ananassa Duch.) under Protected Conditions. J. Appl. Nat. Sci. 2017, 9, 1676–1681. [Google Scholar] [CrossRef]
- Saha, T.; Ghosh, B.; Debnath, S.; Kundu, S.; Bhattacharjee, A. Effect of Plant Growth Regulators on Growth, Yield and Quality of Strawberry (Fragaria × Ananassa Duch.) Cv. Winter Dawn in the Gangetic Alluvial Region of West Bengal, India. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1706–1712. [Google Scholar] [CrossRef]
- Dong, Y.; Song, M.; Liu, X.; Tian, R.; Zhang, L.; Gan, L. Effects of Exogenous KT and BA on Fruit Quality in Strawberry (Fragaria vesca). J. Hortic. Sci. Biotechnol. 2022, 97, 236–243. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Xiao, J.; Guo, G.; Jeong, B.R. Foliar Thidiazuron Promotes the Growth of Axillary Buds in Strawberry. Agronomy 2021, 11, 594. [Google Scholar] [CrossRef]
- Ahmed, A.; Al-Madhagi, I.A.H.; Hasen, S.M.Z.; Zain, A.M.; Yousef, W.A. The Influence of Exogenous Hormone on the Fruit Quality of Strawberry (Fragaria × Ananassa Duch). Int. J. Dev. Sustain. 2017, 6, 1250–1257. [Google Scholar]
- Kour, S.; Kumar, R.; Wali, V.K.; Sharma, A.; Bakshi, P. Impact of Benzyladenine and Gibberellic Acid on Quality and Economics of Runner Production in Chandler Strawberry (Fragaria × Ananassa) under Subtropical Climate. Indian J. Agric. Sci. 2017, 87, 112–115. [Google Scholar] [CrossRef]
- Yoshida, S.; Mandel, T.; Kuhlemeier, C. Stem Cell Activation by Light Guides Plant Organogenesis. Genes Dev. 2011, 25, 1439–1450. [Google Scholar] [CrossRef]
- Di Marzo, M.; Herrera-Ubaldo, H.; Caporali, E.; Novák, O.; Strnad, M.; Balanzà, V.; Ezquer, I.; Mendes, M.A.; de Folter, S.; Colombo, L. SEEDSTICK Controls Arabidopsis Fruit Size by Regulating Cytokinin Levels and FRUITFULL. Cell Rep. 2020, 30, 2846–2857. [Google Scholar] [CrossRef]
- Skalák, J.; Vercruyssen, L.; Claeys, H.; Hradilová, J.; Cermy, M.; Novák, O.; Placková, L.; Saiz-Fernández, I.; Skaláková, P.; Coppens, F.; et al. Multifaced Activity of Cytokinin in Leaf Development Shapes Its Size and Structure in Arabiopsis. Plant J. 2019, 97, 803–804. [Google Scholar] [CrossRef]
- Li, Y.; Feng, J.; Cheng, L.; Dai, C.; Gao, Q.; Liu, Z.; Kang, C. Gene Expression Profiling of the Shoot Meristematic Tissues in Woodland Strawberry Fragaria vesca. Front. Plant Sci. 2019, 10, 1624. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Dobrev, P.I.; Hoyerová, K.; Petrášek, J. Analytical Determination of Auxins and Cytokinins. Methods Mol. Biol. 2017, 1569, 31–39. [Google Scholar]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant Regulation of Meristem Identity and Plant Architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 Years of GDR: New Data and Functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. Available online: https://www.rosaceae.org/ (accessed on 10 June 2022). [CrossRef] [PubMed]








| Biosynthesis * | Signaling | ||||||
|---|---|---|---|---|---|---|---|
| Isopentenyl transferase | Lonely guy | Histidine Kinase | Histidine Phospho transferase | ||||
| Identifier | IPT | Identifier | LOG | Identifier | HK | Identifier | HP |
| FVE03725 | FvIPT1 | FVE16128 | FvLOG1 | FVE14583 | FvHK1 | FVE06646 | FvHP1 |
| FVE23509 | FvIPT2 | FVE30641 | FvLOG2 | FVE21593 | FvHK2 | FVE27751 | FvHP2 |
| FVE30601 | FvIPT3 | FVE09744 | FvLOG3 | FVE30742 | FvHK3 | FVE09006 | FvHP3 |
| FVE27343 | FvIPT4 | FVE15306 | FvLOG4 | FVE04136 | FvHK4 | FVE25730 | FvHP4 |
| FVE27842 | FvIPT5 | FVE08500 | FvLOG5 | FVE22351 | FvHP5 | ||
| FVE06080 | FvIPT6 | FVE02714 | FvLOG6 | FVE15747 | FvHP6 | ||
| FVE27180 | FvIPT7 | FVE30448 | FvLOG7 | FVE25334 | FvHP7 | ||
| FVE30673 | FvLOG8 | FVE30249 | FvHP8 | ||||
| FVE30477 | FvLOG9 | FVE11292 | FvHP9 | ||||
| Signaling * | Degradation * | ||||||
| RR type A | RR type B | RR type C | Cytokinin Oxidase/Dehydrogenase | ||||
| Identifier | RRA | Identifier | RRB | Identifier | RRC | Identifier | CKX |
| FVE00726 | FvRRA1 | FVE06245 | FvRRB8 | FVE08408 | FvRRC15 | FVE14452 | FvCKX1 |
| FVE09501 | FvRRA2 | FVE06943 | FvRRB9 | FVE08409 | FvRRC16 | FVE30654 | FvCKX2 |
| FVE11196 | FvRRA3 | FVE05178 | FvRRB10 | FVE16714 | FvRRC17 | FVE21961 | FvCKX3 |
| FVE14296 | FvRRA4 | FVE10041 | FvRRB11 | FVE08468 | FvRRC18 | FVE12649 | FvCKX4 |
| FVE21358 | FvRRA5 | FVE13325 | FvRRB12 | FVE12648 | FvCKX5 | ||
| FVE21885 | FvRRA6 | FVE15029 | FvRRB13 | FVE15382 | FvCKX6 | ||
| FVE28096 | FvRRA7 | FVE15958 | FvRRB14 | FVE30442 | FvCKX7 | ||
| FVE15204 | FvCKX8 | ||||||
| Fall–Winter Experiment (December 2020–February 2021) | Spring–Summer Experiment (April–July 2021) | ||||
|---|---|---|---|---|---|
| Treatment | Concentration ppm | Frequency of Application | Treatment | Concentration ppm | Frequency of Application |
| 1 | Control | 1 | Control | ||
| 2 | 50 | 1 | 2 | 50 | 1 |
| 3 | 50 | 2 | 3 | 50 | 3 |
| 4 | 50 | 3 | 4 | 100 | 1 |
| 5 | 50 | 4 | |||
| 6 | 50 | 2 | |||
| 7 | 50 | 1 | |||
| F. vesca | F. × ananassa Blast in GDR | |||||
|---|---|---|---|---|---|---|
| Gene | ID | CDS | ID | Length pb | Identity | % |
| CKX2 | FvH4_3g03260 | 1596 | FxaC_9g52120.t1 FxaC_11g00140.t1 | 1578 1584 | 1576/1596 1548/1602 | 98.75 96.63 |
| CKX3 | FvH4_6g24620 | 1638 | FxaC_21g36910.t1 FxaC_23g15950.t1 | 1638 1626 | 1634/1638 1609/1638 | 99.76 98.23 |
| CXK7 | FvH4_3g04610 | 1482 | FxaC_9g50630.t1 FxaC_12g45970.t1 FxaC_10g02600.t1 | 1482 1809 1543 | 1472/1482 1424/1482 1420/1482 | 99.33 96.09 95.82 |
| IPT2 | FvH4_3g29650 | 1446 | FxaC_12g15640.t1 FxaC_11g30460.t1 | 1857 1813 | 1425/1449 1420/1446 | 98.34 98.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Rojas, M.; Díaz-Ramírez, D.; Ortíz-Ramírez, C.I.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M.; Ferrándiz, C.; Abraham-Juárez, M.d.R.; Marsch-Martínez, N. The Role of Cytokinins during the Development of Strawberry Flowers and Receptacles. Plants 2023, 12, 3672. https://doi.org/10.3390/plants12213672
Pérez-Rojas M, Díaz-Ramírez D, Ortíz-Ramírez CI, Galaz-Ávalos RM, Loyola-Vargas VM, Ferrándiz C, Abraham-Juárez MdR, Marsch-Martínez N. The Role of Cytokinins during the Development of Strawberry Flowers and Receptacles. Plants. 2023; 12(21):3672. https://doi.org/10.3390/plants12213672
Chicago/Turabian StylePérez-Rojas, Moises, David Díaz-Ramírez, Clara Inés Ortíz-Ramírez, Rosa M. Galaz-Ávalos, Víctor M. Loyola-Vargas, Cristina Ferrándiz, Ma. del Rosario Abraham-Juárez, and Nayelli Marsch-Martínez. 2023. "The Role of Cytokinins during the Development of Strawberry Flowers and Receptacles" Plants 12, no. 21: 3672. https://doi.org/10.3390/plants12213672
APA StylePérez-Rojas, M., Díaz-Ramírez, D., Ortíz-Ramírez, C. I., Galaz-Ávalos, R. M., Loyola-Vargas, V. M., Ferrándiz, C., Abraham-Juárez, M. d. R., & Marsch-Martínez, N. (2023). The Role of Cytokinins during the Development of Strawberry Flowers and Receptacles. Plants, 12(21), 3672. https://doi.org/10.3390/plants12213672








