Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia
Abstract
1. Introduction
2. Results
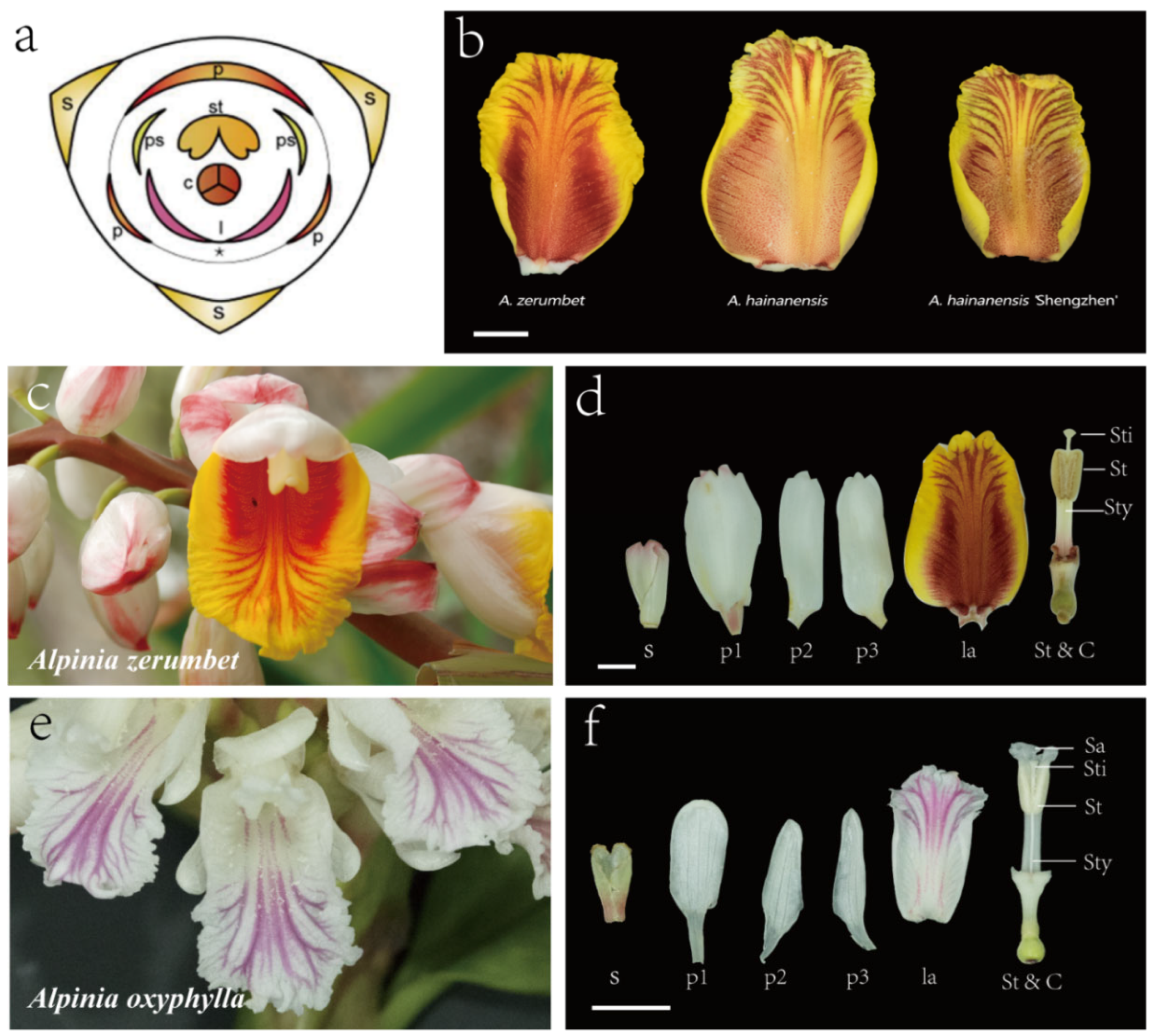
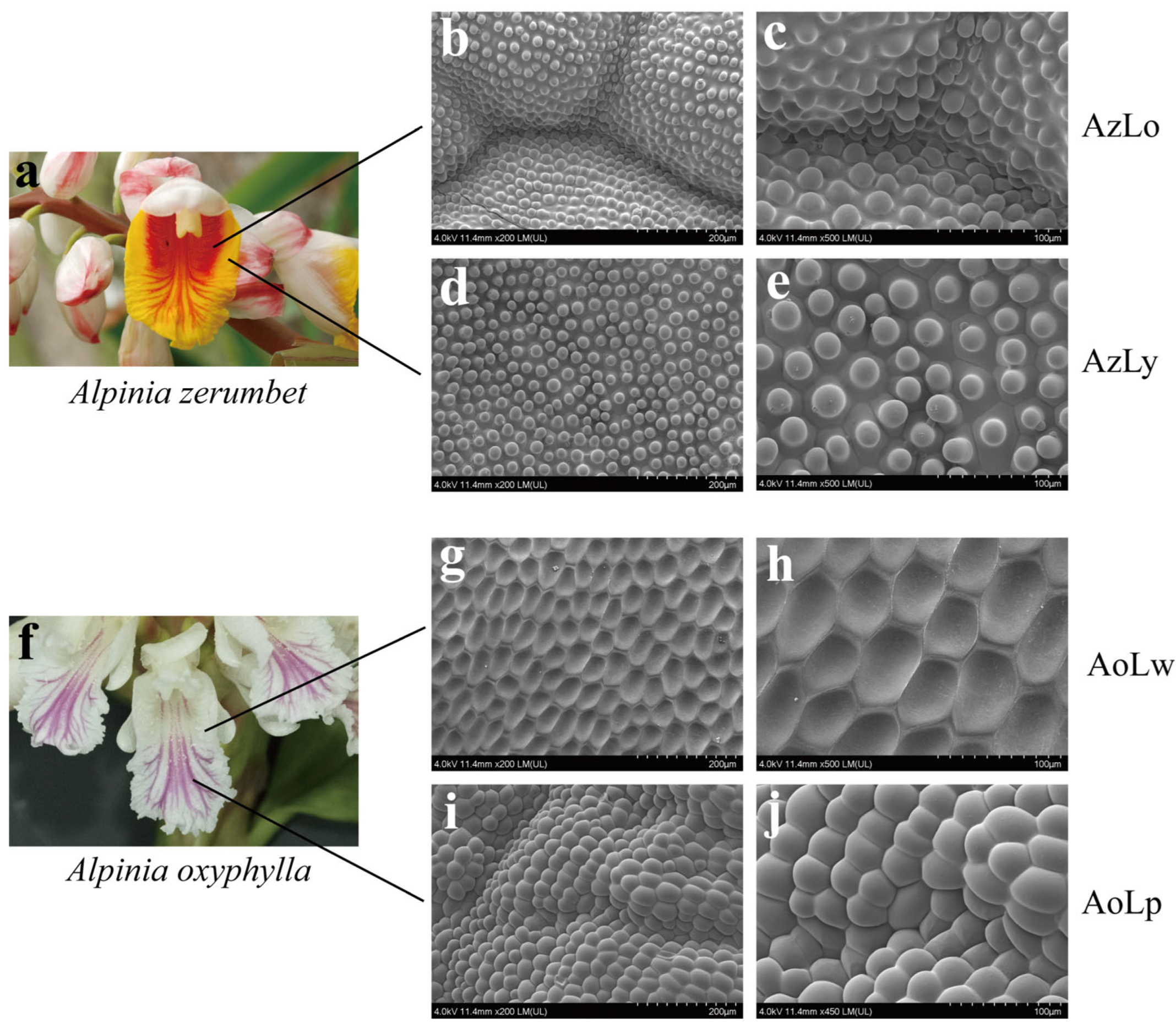
2.1. Differences in Epidermal Cell Morphology in Four Pigmented Segments of Alpinia Labellum
2.2. Flavonoid Metabolic Differences in the Four Pigmented Segments of Alpinia Labella
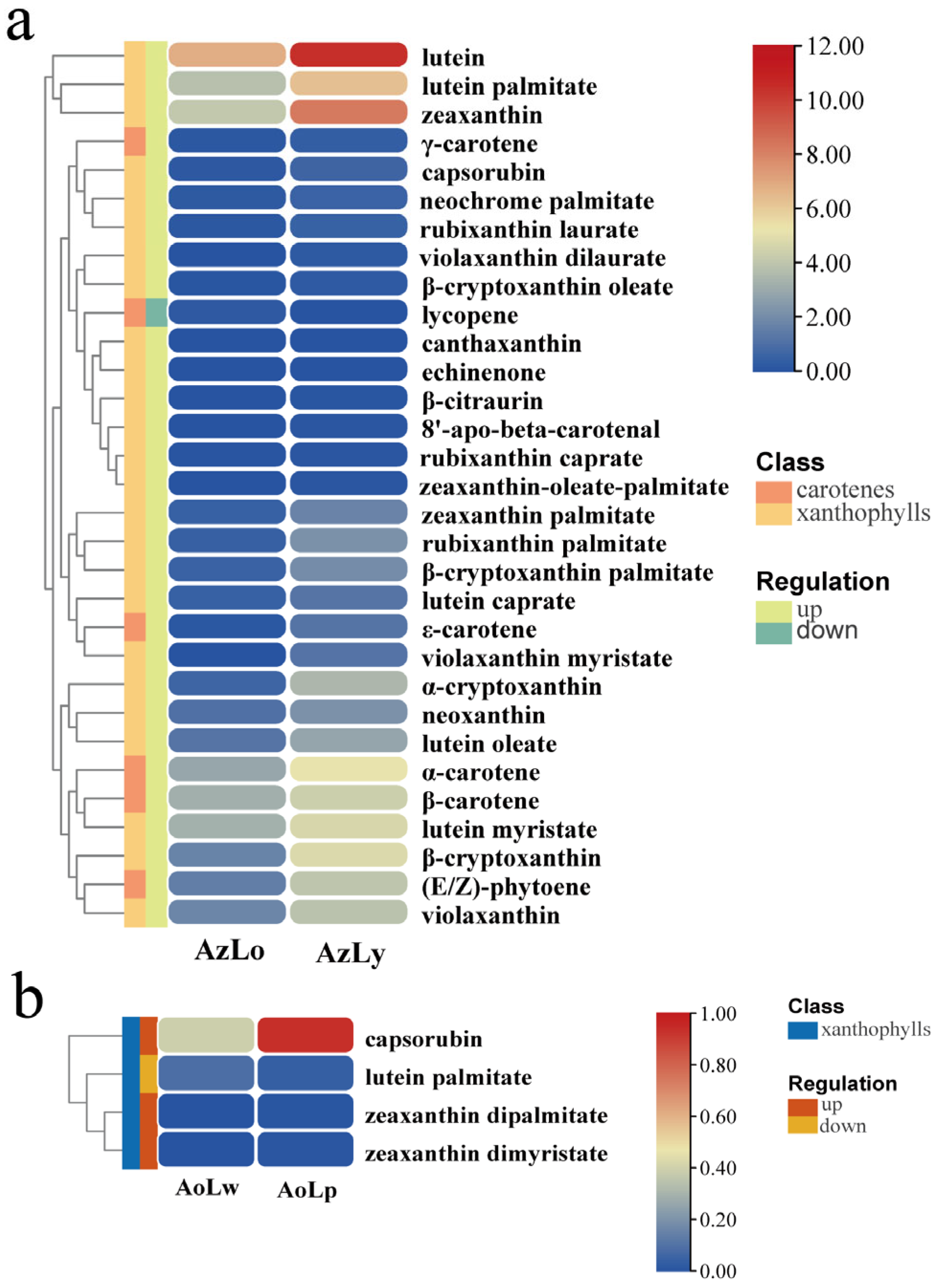
2.3. Carotenoid Metabolic Differences in the Four Pigmented Segments of Alpinia Labella
2.4. Transcriptome Sequencing and Unigene Functional Annotation
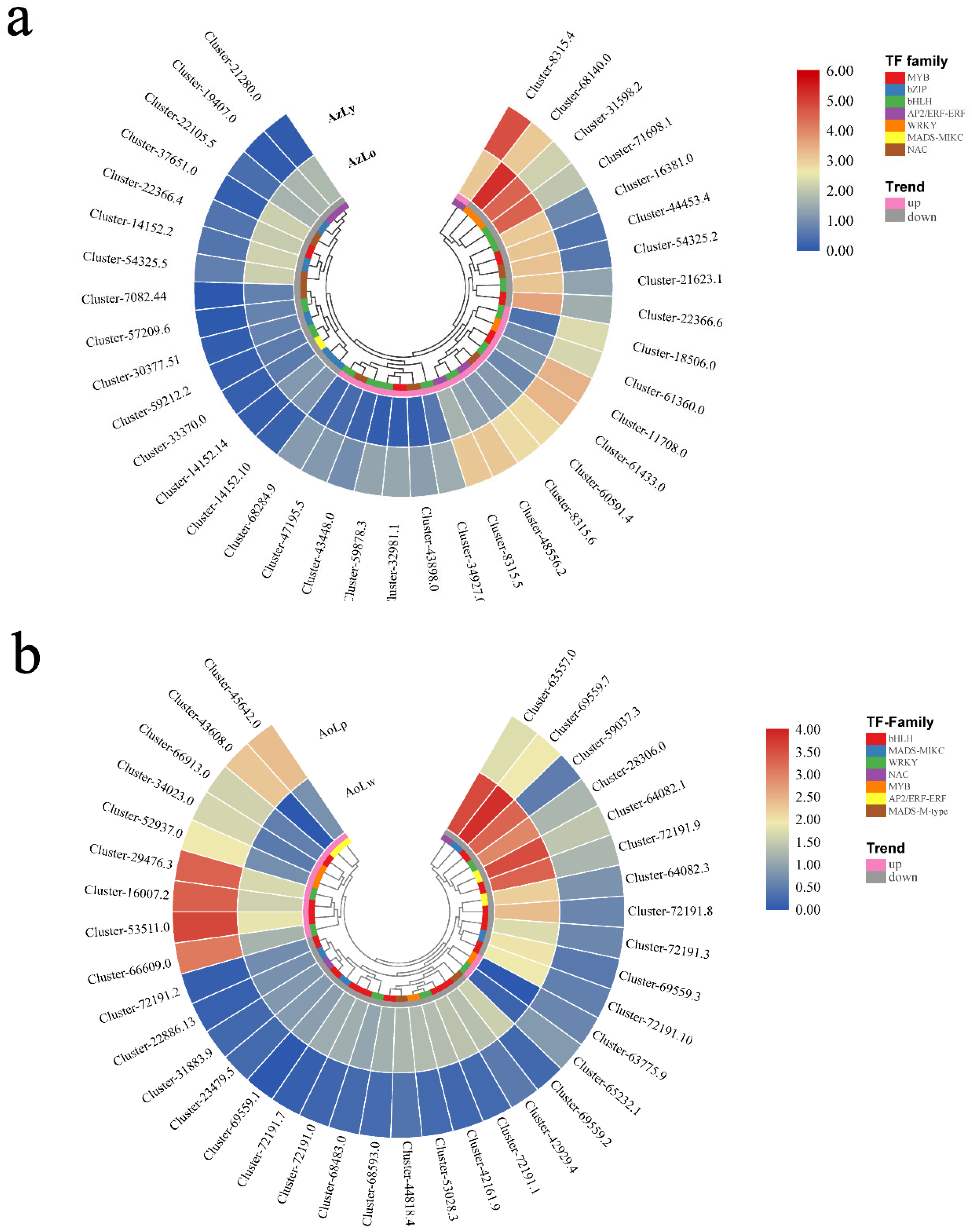
2.5. Differentially Expressed Genes (DEGs) Involved in Flavonoid and Carotenoid Biosynthesis
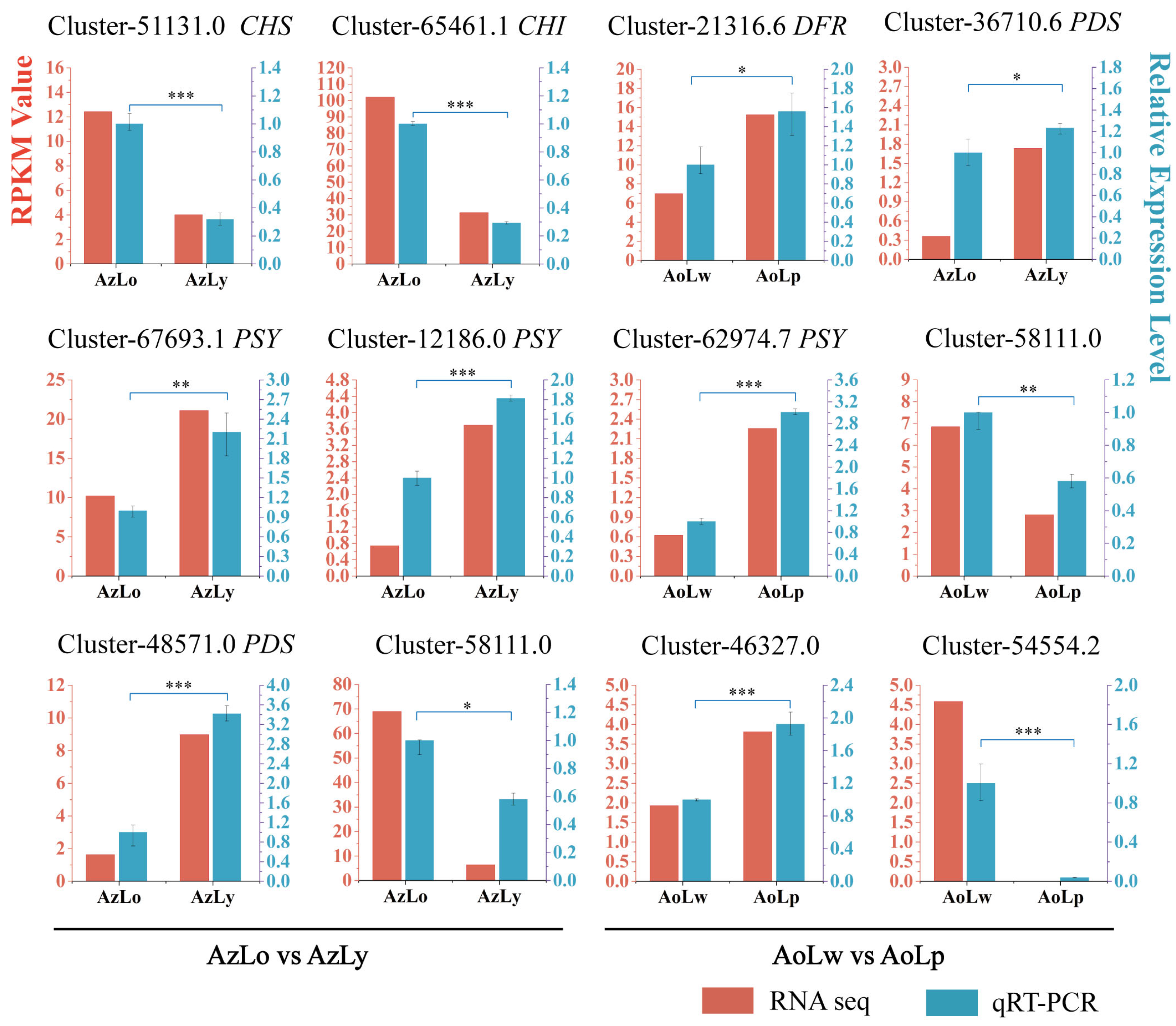
2.6. Validation of Key Pigmentation-Related Gene Expression
3. Discussion
3.1. Possible Correlation between the Various Flower Color Displays of Alpinia Labellum with the Morphology of Individual Epidermal Cells and Cell Populations
3.2. The Differential Accumulation of Flavonoids and Anthocyanins in Different Color Segments Has a Significant Impact on the Color Formation of Alpinia Labella
3.3. Carotenoid Biosynthesis Mainly Contributes to the Yellow Pigmentation of Alpinia Labellum
4. Materials and Methods
4.1. Plant Materials
4.2. Cryo-Scanning Electron Microscopy
4.3. Transcriptome Sequencing
4.4. Gene Functional Annotation and Differential Expression
4.5. Real-Time Quantitative PCR (qRT-PCR) for RNA Validation
4.6. Flavonoid Metabolite Isolation and Analysis
4.7. Carotenoids, Metabolite Isolation and Analysis
4.8. Selection of Differential Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahlgren, R.M.T.; Rasmussen, F. Monocotyledon evolution: Characters and phylogenetic estimation. Evol. Biol. 1983, 16, 255–395. [Google Scholar]
- Kirchoff, B.K. Floral ontogeny and evolution in the ginger group of the Zingiberales. In Aspects of Floral Development; Leins, P., Tucker, S.C., Eds.; Cramer: Berlin, Germany, 1988; pp. 45–56. [Google Scholar]
- Kress, W.J. The Phylogeny and classification of the Zingiberales. Ann. Mo. Bot. Gard. 1990, 77, 698–721. [Google Scholar] [CrossRef]
- Kirchoff, B.K. Inflorescence and flower development in the Hedychieae (Zingiberaceae): Hedychium. Can. J. Bot. 1997, 75, 581–594. [Google Scholar] [CrossRef][Green Version]
- Kirchoff, B.K.; Lagomarsino, L.P.; Newman, W.H.; Bartlett, M.E.; Specht, C.D. Early floral development of Heliconia latispatha (Heliconiaceae), a key taxon for understanding the evolution of flower development in the Zingiberales. Am. J. Bot. 2009, 96, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower coloration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Yuan, Y.-W.; Rebocho, A.B.; Sagawa, J.M.; Stanley, L.E.; Bradshaw, H.D. Competition between anthocyanin and flavonol biosynthesis produces spatial pattern variation of floral pigments between Mimulus species. Proc. Natl. Acad. Sci. USA 2016, 113, 2448. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of novel flower colors in floricultural plants: Recent advances via transgenic approaches. Methods Mol. Biol. 2010, 589, 325–347. [Google Scholar]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef]
- Shang, Y.; Venail, J.; Mackay, S.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Martin, C.R.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef]
- Yasuda, H. Studies on “Bluing Effect” in the Petals of Red Rose. Cytologia 1974, 39, 107–112. [Google Scholar] [CrossRef]
- Mudalige, R.; Kuehnle, A.; Amore, T. Pigment distribution and epidermal cell shape in Dendrobium species and hybrids. HortScience 2003, 38, 573–577. [Google Scholar] [CrossRef]
- Yin, X.; Lin, X.; Liu, Y.; Irfan, M.; Chen, L.; Zhang, L. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in diverse petal tissues in the lily cultivar ‘Vivian’. BMC Plant Biol. 2020, 20, 446. [Google Scholar] [CrossRef] [PubMed]
- Quintana, A.; Albrechtová, J.; Griesbach, R.J.; Freyre, R. Anatomical and biochemical studies of anthocyanidins in flowers of Anagallis monelli L. (Primulaceae) hybrids. Sci. Hortic. 2007, 112, 413–421. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid metabolism in plants: The role of plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [PubMed]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001, 4, 210–218. [Google Scholar] [CrossRef]
- Han, Y.; Wang, X.; Chen, W.; Dong, M.; Yuan, W.; Liu, X.; Shang, F. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans. Tree Genet. Genomes 2014, 10, 329–338. [Google Scholar] [CrossRef]
- Moehs, C.P.; Tian, L.; Osteryoung, K.W.; Dellapenna, D. Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol. Biol. 2001, 45, 281–293. [Google Scholar] [CrossRef]
- Luo, X.; Sun, D.; Wang, S.; Luo, S.; Fu, Y.; Niu, L.; Shi, Q.; Zhang, Y. Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flowers. Hortic. Res. 2021, 1, 008. [Google Scholar]
- Zhao, T.; Specht, C.D.; Dong, Z.; Ye, Y.; Liu, H.; Liao, J. Transcriptome analyses provide insights into development of the Zingiber zerumbet flower, revealing potential genes related to floral organ formation and patterning. Plant Growth Regul. 2020, 90, 331–345. [Google Scholar] [CrossRef]
- Zhao, T.; Piñeyro-Nelson, A.; Yu, Q.; Pan, X.; Hu, X.; Liu, H.; Liao, J. Transcriptome-wide analyses provide insights into development of the Hedychium Coronarium flower, revealing potential roles of PTL. J. Plant Biol. 2021, 64, 431–445. [Google Scholar] [CrossRef]
- Kay, Q.O.N.; Daoud, H.S.; Stirton, C.H. Pigment distribution, light reflection and cell structure in petals. Bot. J. Linn. Soc. 1981, 83, 57–83. [Google Scholar] [CrossRef]
- Whitney, H.M.; Bennett, K.V.; Dorling, M.; Sandbach, L.; Prince, D.; Chittka, L.; Glover, B.J. Why do so many petals have conical epidermal cells? Ann. Bot. 2011, 108, 609–616. [Google Scholar] [CrossRef]
- Rudall, P.J. Colorful cones: How did flower color first evolve? J. Exp. Bot. 2020, 71, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Noda, K.; Glover, B.J.; Linstead, P.; Martin, C. Flower color intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 1994, 369, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Dyer, A.G.; Whitney, H.M.; Arnold, S.E.; Glover, B.J.; Chittka, L. Mutations perturbing petal cell shape and anthocyanin synthesis influence bumblebee perception of Antirrhinum majus flower color. Arthropod Plant Interact. 2007, 1, 45–55. [Google Scholar] [CrossRef]
- Yang, Q.; Yuan, H.H.; Sun, X.B. Preliminary studies on the changes of flower color during the flowering period in two tree peony cultivars. Acta Hortic. Sin. 2015, 42, 930–938. [Google Scholar]
- Guo, L.; Wang, Y.; da Silva, J.A.T.; Fan, Y.; Yu, X. Transcriptome and chemical analysis reveal putative genes involved in flower color change in Paeonia ‘coral sunset’. Plant Physiol. Bio Chem 2019, 138, 130–139. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Fu, L.; Li, H.; Li, L.; Yu, H.; Wang, L. Reason of flower color change in Lonicera japonica. Sci. Silvae Sin. 2013, 49, 155–161. (In Chinese) [Google Scholar]
- Xia, Y.; Chen, W.; Xiang, W.; Wang, D.; Xue, B.; Liu, X.; Xing, L.; Wu, D.; Wang, S.; Guo, Q.; et al. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in Lonicera japonica flower petals during color-transition. BMC Plant Biol. 2021, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Truesdale, M.R.; Bird, C.R.; Schuch, W.; Bramley, P.M. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol. 1994, 105, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Moise, A.R.; Al-Babili, S.; Wurtzel, E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014, 114, 164–193. [Google Scholar] [PubMed]
- Ducreux, L.J.; Morris, W.L.; Hedley, P.E.; Shepherd, T.; Davies, H.V.; Millam, S.; Taylor, M.A. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. J. Exp. Bot. 2004, 56, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, M.; Kishimoto, S.; Nakayama, M. Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 2010, 129, 100–107. [Google Scholar] [CrossRef]
- He, J.; Jiao, Y. Next-generation sequencing applied to flower development: RNA-seq. In Flower Development: Methods and Protocols; Riechmann, J.L., Wellmer, F., Eds.; Springer: New York, NY, USA, 2014; Volume 1110, pp. 401–411. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Krinsky, N.I.; Mayne, S.T.; Sies, H. Carotenoids in Health and Disease; CRC Press Inc.: Boca Raton, FL, USA, 2004; pp. 1–30. [Google Scholar]
- Bartleyg, E.; Scolnik, P.A. Plant carotenoids: Pigments for photoprotection, visual attraction and human health. Plant Cell 1995, 7, 1027–1038. [Google Scholar]
- Inbaraj, B.S.; Lu, H.; Hung, C.F.; Wu, W.B.; Lin, C.L.; Chen, B.H. Determination of carotenoids and their esters in fruits of Lycium barbarum Linnaeus by HPLC-DAD-APCI-MS. J. Pharm. Biomed. Anal. 2008, 47, 812–818. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Yu, Q.; Lin, C.; Liu, H.; Dong, L.; Feng, X.; Liao, J. Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia. Plants 2023, 12, 3766. https://doi.org/10.3390/plants12213766
Zhao T, Yu Q, Lin C, Liu H, Dong L, Feng X, Liao J. Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia. Plants. 2023; 12(21):3766. https://doi.org/10.3390/plants12213766
Chicago/Turabian StyleZhao, Tong, Qianxia Yu, Canjia Lin, Huanfang Liu, Limei Dong, Xinxin Feng, and Jingping Liao. 2023. "Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia" Plants 12, no. 21: 3766. https://doi.org/10.3390/plants12213766
APA StyleZhao, T., Yu, Q., Lin, C., Liu, H., Dong, L., Feng, X., & Liao, J. (2023). Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia. Plants, 12(21), 3766. https://doi.org/10.3390/plants12213766




