The Underexplored Mechanisms of Wheat Resistance to Leaf Rust
Abstract
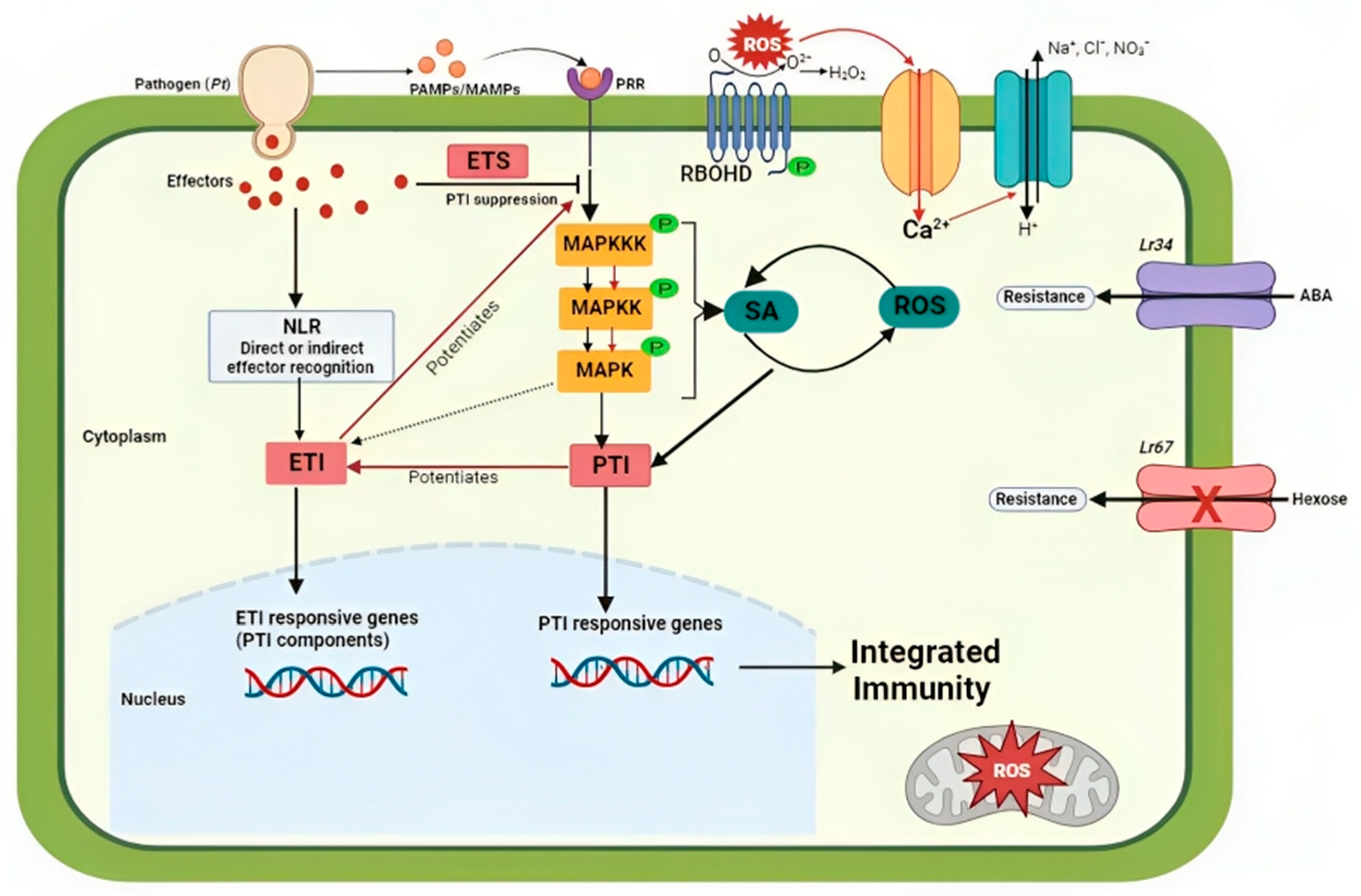
:1. Introduction
2. Host Range, Pathogenic Profile, and Evolutionary Basis
2.1. Host Range and Pathogenic Profile
2.2. Evolutionary Basis of Host–Pathogen Interactions
3. Wheat Infection by Pt
3.1. Penetration and Colonization of Wheat by Pt
3.2. Molecular Basis Underlying Wheat–Pt Interactions
4. Mechanisms of Wheat Resistance to Leaf Rust
4.1. Cloning and Characterization of Wheat Lr Genes
4.2. Prehaustorial Resistance
4.3. Posthaustorial Resistance
4.4. Systemic Acquired Resistance
5. ROS and Their Role in Wheat Defense against Leaf Rust
6. Interplay between Climatic Factors, Genetic Characteristics, Planting Dates, and Disease Dynamics
7. Future Prospects and Limitations
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ellinger, D.; Naumann, M.; Falter, C.; Zwikowics, C.; Jamrow, T.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013, 161, 1433–1444. [Google Scholar] [CrossRef]
- Thordal-Christensen, H. Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 2003, 6, 351–357. [Google Scholar] [CrossRef]
- Bolton, M.D.; Kolmer, J.A.; Garvin, D.F. Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 2008, 9, 563–575. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Delfan, S.; Bihamta, M.R.; Dadrezaei, S.T.; Abbasi, A.; Alipour, H. Identification sources of resistance for leaf rust (Puccinia triticina Eriks.) in Iranian wheat germplasm. Iran. J. Plant Prot. Sci. 2022, 52, 115–133. [Google Scholar]
- Delfan, S.; Bihamta, M.R.; Dadrezaei, S.T.; Abbasi, A.; Alipoor, H. Exploring genomic regions involved in bread wheat resistance to leaf rust at seedling/adult stages by using GWAS analysis. BMC Genom. 2023, 24, 83. [Google Scholar] [CrossRef]
- Talebi, R.; Mahboubi, M.; Naji, A.M.; Mehrabi, R. Physiological specialization of Puccinia triticina and genome-wide association mapping provide insights into the genetics of wheat leaf rust resistance in Iran. Sci. Rep. 2023, 13, 4398. [Google Scholar] [CrossRef]
- Huerta-Espino, J.; Singh, R.P.; Germán, S.; McCallum, B.D.; Park, R.F.; Chen, W.Q.; Bhardwaj, S.C.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- McCallum, B.D.; Hiebert, C.W.; Cloutier, S.; Bakkeren, G.; Rosa, S.B.; Humphreys, D.G.; Marais, G.F.; McCartney, C.A.; Panwar, V.; Rampitsch, C. A review of wheat leaf rust research and the development of resistant cultivars in Canada. Can. J. Plant Pathol. 2016, 38, 1–18. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Pélissier, R.; Violle, C.; Morel, J.-B. Plant immunity: Good fences make good neighbors? Curr. Opin. Plant Biol. 2021, 62, 102045. [Google Scholar] [CrossRef]
- Scheres, B.; van der Putten, W.H. The plant perceptron connects environment to development. Nature 2017, 543, 337–345. [Google Scholar] [CrossRef]
- Nobori, T.; Tsuda, K. The plant immune system in heterogeneous environments. Curr. Opin. Plant Biol. 2019, 50, 58–66. [Google Scholar] [CrossRef]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef]
- Singh, S.; Awasthi, L.P.; Verma, H.N. Systemic resistance inducers from plants—An ecofriendly approach for the management of viral diseases of crops. In Applied Plant Virology; Awasthi, L.P., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 603–617. [Google Scholar]
- Riar, A.K.; Chhuneja, P.; Keller, B.; Singh, K. Mechanism of leaf rust resistance in wheat wild relatives, Triticum monococcum L. and T. boeoticum L. Plant Genet. Resour. 2021, 19, 320–327. [Google Scholar] [CrossRef]
- Heath, M.C. A generalized concept of host-parasite specificity. Phytopathology 1981, 71, 1121–1123. [Google Scholar] [CrossRef]
- Niks, R.E.; Dekens, R.G. Prehaustorial and post-haustorial resistance to wheat leaf rust in diploid wheat seedlings. Phytopathology 1991, 81, 847–851. [Google Scholar] [CrossRef]
- Wei, Z.Z.; Klymiuk, V.; Bocharova, V.; Pozniak, C.; Fahima, T. A Post-Haustorial Defense Mechanism is Mediated by the Powdery Mildew Resistance Gene, PmG3M, Derived from Wild Emmer Wheat. Pathogens 2020, 9, 418. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, H.; Yao, J.; Wang, X.; Xu, J.; Han, Q.; Wei, G.; Huang, L.; Kang, Z. Characterization of non-host resistance in broad bean to the wheat stripe rust pathogen. BMC Plant Biol. 2012, 12, 96. [Google Scholar] [CrossRef]
- Jacobs, T. Haustorium Formation and Cell Wall Appositions in Susceptible and Partially Resistant Wheat and Barley Seedlings Infected with Wheat Leaf Rust. J. Phytopathol. 1989, 127, 250–261. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Faoro, F. Induced systemic resistance against systemic viruses: A feasible approach? IOBC-WPRS Bull. 2013, 89, 199–203. [Google Scholar]
- Averyanov, A. Oxidative burst and plant disease resistance. Front. Biosci. (Elite Ed.) 2009, 1, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal 2009, 2, ra45. [Google Scholar] [CrossRef] [PubMed]
- Roelfs, A.P.; Singh, R.P.; Saari, E.E. Rust Diseases of Wheat. Concepts and Methods of Disease Management; CIMMYT: Mexico City, Mexico, 1992. [Google Scholar]
- Mebrate, S.A.; Oerke, E.C.; Dehne, H.W.; Pillen, K. Mapping of the leaf rust resistance gene Lr38 on wheat chromosome arm 6DL using SSR markers. Euphytica 2008, 162, 457–466. [Google Scholar] [CrossRef]
- Abebe, W. Wheat Leaf Rust Disease Management: A Review. J. Plant Pathol. Microbiol. 2021, 12, 1–8. [Google Scholar]
- Kolmer, J.A. Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 2005, 8, 441–449. [Google Scholar] [CrossRef]
- Anikster, Y.; Eilam, T.; Bushnell, W.R.; Kosman, E. Spore dimensions of Puccinia species of cereal hosts as determined by image analysis. Mycologia 2005, 97, 474–484. [Google Scholar] [CrossRef]
- Campbell, F.T. The science of risk assessment for phytosanitary regulation and the impact of changing trade regulations: The approach to phytosanitary safeguards mandated by the World Trade Organization may hinder adoption of the most efficient methods to protect ecosystems from introductions of invasive species. BioScience 2001, 51, 148–153. [Google Scholar]
- Perrings, C.; Williamson, M.; Barbier, E.B.; Delfino, D.; Dalmazzone, S.; Shogren, J.; Simmons, P.; Watkinson, A. Biological invasion risks and the public good: An economic perspective. Conserv. Ecol. 2002, 6, 1. [Google Scholar] [CrossRef]
- Brown, J.K.M.; Hovmøller, M.S. Aerial dispersal of fungi on the global and continental scales and its consequences for plant disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef]
- Ali, S.; Gladieux, P.; Leconte, M.; Gautier, A.; Justesen, A.F.; Hovmøller, M.S.; Enjalbert, J.; de Vallavieille-Pope, C. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f. sp. tritici. PLoS Pathog. 2014, 10, e1003903. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Thompson, J.N. The evolution of species interactions. Science 1999, 284, 2116–2118. [Google Scholar] [CrossRef]
- Thompson, J.N. The Geographic Mosaic of Coevolution; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Burdon, J.J.; Thrall, P.H. What have we learned from studies of wild plant-pathogen associations?—The dynamic interplay of time, space and life-history. Eur. J. Plant Pathol. 2014, 138, 417–429. [Google Scholar] [CrossRef]
- Upson, J.L.; Zess, E.K.; Białas, A.; Wu, C.H.; Kamoun, S. The coming of age of EvoMPMI: Evolutionary molecular plant-microbe interactions across multiple timescales. Curr. Opin. Plant Biol. 2018, 44, 108–116. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Valè, G.; Mastrangelo, A.M. Regulation and Evolution of NLR Genes: A Close Interconnection for Plant Immunity. Int. J. Mol. Sci. 2018, 19, 1662. [Google Scholar] [CrossRef]
- de Vries, S.; Stukenbrock, E.H.; Rose, L.E. Rapid evolution in plant-microbe interactions—An evolutionary genomics perspective. New Phytol. 2020, 226, 1256–1262. [Google Scholar] [CrossRef]
- Van, V. A new evolutionary law. Evol. Theory 1973, 1, 284–314. [Google Scholar]
- Brockhurst, M.A.; Chapman, T.; King, K.C.; Mank, J.E.; Paterson, S.; Hurst, G.D. Running with the Red Queen: The role of biotic conflicts in evolution. Proc. Biol. Sci. 2014, 281, 20141382. [Google Scholar] [CrossRef]
- Sironi, M.; Cagliani, R.; Forni, D.; Clerici, M. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015, 16, 224–236. [Google Scholar] [CrossRef]
- Han, G.-Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Webster, J.P.; Domingo, E.; Charlesworth, B.; Levin, B.R. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002, 32, 569–577. [Google Scholar] [CrossRef]
- Pfennig, K.S. Evolution of pathogen virulence: The role of variation in host phenotype. Proc. Biol. Sci. 2001, 268, 755–760. [Google Scholar] [CrossRef]
- Brown, J.K.; Tellier, A. Plant-parasite coevolution: Bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 2011, 49, 345–367. [Google Scholar] [CrossRef]
- Wynn, W.K. Appressorium Formation over Stomates by the Bean Rust Fungus: Response to a Surface Contact Stimulus. Phytopathology 1976, 66, 136–146. [Google Scholar] [CrossRef]
- Wynn, W.K.; Staples, R.C. Tropisms of fungi in host recognition. In Plant Disease Control: Resistance and Susceptibility; Staples, R.C., Toenniessen, G.H., Eds.; John Wiley and Sons: New York, NY, USA, 1981; p. 45. [Google Scholar]
- Niks, R.E. Effect of germ tube length on the fate of sporelings of Puccinia hordei in susceptible and resistant barley. Phytopathology 1990, 80, 57–60. [Google Scholar] [CrossRef]
- Bruce, M.; Neugebauer, K.; Joly, D.; Migeon, P.; Cuomo, C.; Wang, S.; Akhunov, E.; Bakkeren, G.; Kolmer, J.; Fellers, J. Using transcription of six Puccinia triticina races to identify the effective secretome during infection of wheat. Front. Plant Sci. 2014, 4, 520. [Google Scholar] [CrossRef]
- Kiran, K.; Rawal, H.C.; Dubey, H.; Jaswal, R.; Devanna, B.N.; Gupta, D.K.; Bhardwaj, S.C.; Prasad, P.; Pal, D.; Chhuneja, P.; et al. Draft Genome of the Wheat Rust Pathogen (Puccinia triticina) Unravels Genome-Wide Structural Variations during Evolution. Genome Biol. Evol. 2016, 8, 2702–2721. [Google Scholar] [CrossRef]
- Cui, Z.; Wu, W.; Fan, F.; Wang, F.; Liu, D.; Di, D.; Wang, H. Transcriptome analysis of Lr19-virulent mutants provides clues for the AvrLr19 of Puccinia triticina. Front. Microbiol. 2023, 14, 1062548. [Google Scholar] [CrossRef]
- Qi, Y.; Wei, J.; Zhang, N.; Yang, W.; Liu, D. Puccinia triticina effector protein Pt18906 triggered two-layer defense reaction in TcLr27+31. Sci. Agric. Sin. 2020, 53, 2371–2384. [Google Scholar] [CrossRef]
- Qi, Y.; Li, J.; Mapuranga, J.; Zhang, N.; Chang, J.; Shen, Q.; Zhang, Y.; Wei, J.; Cui, L.; Liu, D.; et al. Wheat leaf rust fungus effector Pt13024 is avirulent to TcLr30. Front. Plant Sci. 2022, 13, 1098549. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shen, S.; Cui, Z.; Yuan, S.; Qu, P.; Jia, H.; Meng, L.; Hao, X.; Liu, D.; Ma, L.; et al. Puccinia triticina effector protein Pt_21 interacts with wheat thaumatin-like protein TaTLP1 to inhibit its antifungal activity and suppress wheat apoplast immunity. Crop J. 2023, 11, 1431–1440. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, W.; Liu, D. Characteristics and Function Analysis of Wheat Leaf Rust Effector Protein Pt2567. Master’s Thesis, Agricultural University of Hebei, Baoding, China, 2014. [Google Scholar]
- Segovia, V.; Bruce, M.; Shoup Rupp, J.L.; Huang, L.; Bakkeren, G.; Trick, H.N.; Fellers, J.P. Two small secreted proteins from Puccinia triticina induce reduction of ß-glucoronidase transient expression in wheat isolines containing Lr9, Lr24 and Lr26. Can. J. Plant Pathol. 2016, 38, 91–102. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, J.M. Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 2013, 55, 1271–1286. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, J.; Shan, L.; Meng, X. Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 2018, 131, jcs209353. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.-i.; Yasuda, S. Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Ranf, S.; Eschen-Lippold, L.; Pecher, P.; Lee, J.; Scheel, D. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. For. Cell Mol. Biol. 2011, 68, 100–113. [Google Scholar] [CrossRef]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Scheler, C.; Durner, J.; Astier, J. Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 2013, 16, 534–539. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Wang, X.; Liu, N.; Xu, B.; Peng, Q.; Guo, Z.; Fan, B.; Zhu, C.; Chen, Z. Arabidopsis Endoplasmic Reticulum-Localized UBAC2 Proteins Interact with PAMP-INDUCED COILED-COIL to Regulate Pathogen-Induced Callose Deposition and Plant Immunity. Plant Cell 2019, 31, 153–171. [Google Scholar] [CrossRef]
- Huang, W.; Wang, Y.; Li, X.; Zhang, Y. Biosynthesis and Regulation of Salicylic Acid and N-Hydroxypipecolic Acid in Plant Immunity. Mol. Plant 2020, 13, 31–41. [Google Scholar] [CrossRef]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. Cell 2020, 182, 1093–1108.e18. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network Properties of Robust Immunity in Plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Tena, G. PTI and ETI are one. Nat. Plants 2021, 7, 1527. [Google Scholar] [CrossRef]
- Tian, H.; Chen, S.; Wu, Z.; Ao, K.; Yaghmaiean, H.; Sun, T.; Huang, W.; Xu, F.; Zhang, Y.; Wang, S.; et al. Activation of TIR signaling is required for pattern-triggered immunity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Locci, F.; Wanke, F.; Zhang, L.; Saile, S.C.; Joe, A.; Karelina, D.; Hua, C.; Fröhlich, K.; Wan, W.-L.; et al. The EDS1–PAD4–ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 2021, 598, 495–499. [Google Scholar] [CrossRef]
- Ruiz-Bedoya, T.; Wang, P.W.; Desveaux, D.; Guttman, D.S. Cooperative virulence via the collective action of secreted pathogen effectors. Nat. Microbiol. 2023, 8, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hige, J.; Dangl, J.L. Is localized acquired resistance the mechanism for effector-triggered disease resistance in plants? Nat. Plants 2023, 9, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Staskawicz, B.J. Genetics of Plant-Pathogen Interactions Specifying Plant Disease Resistance. Plant Physiol. 2001, 125, 73. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Arya, P.; Acharya, V. Plant STAND P-loop NTPases: A current perspective of genome distribution, evolution, and function. Mol. Genet. Genom. 2018, 293, 17–31. [Google Scholar] [CrossRef]
- Prasad, P.; Savadi, S.; Bhardwaj, S.C.; Gupta, P.K. The progress of leaf rust research in wheat. Fungal Biol. 2020, 124, 537–550. [Google Scholar] [CrossRef]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- van Wersch, S.; Tian, L.; Hoy, R.; Li, X. Plant NLRs: The Whistleblowers of Plant Immunity. Plant Commun. 2020, 1, 100016. [Google Scholar] [CrossRef] [PubMed]
- Rubiales, D.; Niks, R.E. Histological responses in Hordeum chilense to brown and yellow rust fungi. Plant Pathol. 1992, 41, 611–617. [Google Scholar] [CrossRef]
- Lagudah, E.S. Molecular genetics of race non-specific rust resistance in wheat. Euphytica 2011, 179, 81–91. [Google Scholar] [CrossRef]
- Niks, R.E. Haustorium formation by Puccinia hordei in leaves of hypersensitive, partially resistant, and nonhost plant genotypes. Phytopathology 1983, 73, 64–66. [Google Scholar] [CrossRef]
- Niks, R.E. Non-host plant species as donors for resistance to pathogens with narrow host range.II. Concepts and evidence of non-host resistance. Euphytica 1988, 37, 89–99. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lagudah, E.S.; Spielmeyer, W.; Dodds, P.N. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 2014, 5, 641. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Dinh, H.X.; Singh, D.; Periyannan, S.; Park, R.F.; Pourkheirandish, M. Molecular genetics of leaf rust resistance in wheat and barley. Theor. Appl. Genet. 2020, 133, 2035–2050. [Google Scholar] [CrossRef]
- Andersen, E.J.; Nepal, M.P.; Purintun, J.M.; Nelson, D.; Mermigka, G.; Sarris, P.F. Wheat Disease Resistance Genes and Their Diversification Through Integrated Domain Fusions. Front. Genet. 2020, 11, 898. [Google Scholar] [CrossRef]
- Dinglasan, E.; Periyannan, S.; Hickey, L.T. Harnessing adult-plant resistance genes to deploy durable disease resistance in crops. Essays Biochem. 2022, 66, EBC20210096. [Google Scholar] [CrossRef]
- Kumar, S.; Kamboj, D.; Srivastava, P.; Mishra, C.N.; Singh, G.; Singh, G.P. Broadening Genetic Base of Wheat for Improving Rust Resistance. In New Horizons in Wheat and Barley Research: Global Trends, Breeding and Quality Enhancement; Kashyap, P.L., Gupta, V., Prakash Gupta, O., Sendhil, R., Gopalareddy, K., Jasrotia, P., Singh, G.P., Eds.; Springer: Singapore, 2022; pp. 401–427. [Google Scholar]
- Mapuranga, J.; Zhang, N.; Zhang, L.; Liu, W.; Chang, J.; Yang, W. Harnessing genetic resistance to rusts in wheat and integrated rust management methods to develop more durable resistant cultivars. Front. Plant Sci. 2022, 13, 951095. [Google Scholar] [CrossRef]
- Mao, H.; Jiang, C.; Tang, C.; Nie, X.; Du, L.; Liu, Y.; Cheng, P.; Wu, Y.; Liu, H.; Kang, Z.; et al. Wheat adaptation to environmental stresses under climate change: Molecular basis and genetic improvement. Mol. Plant 2023, 16, 1564–1589. [Google Scholar] [CrossRef]
- Ren, X.; Wang, C.; Ren, Z.; Wang, J.; Zhang, P.; Zhao, S.; Li, M.; Yuan, M.; Yu, X.; Li, Z.; et al. Genetics of Resistance to Leaf Rust in Wheat: An Overview in a Genome-Wide Level. Sustainability 2023, 15, 3247. [Google Scholar] [CrossRef]
- Deblieck, M.; Ordon, F.; Serfling, A. Mapping of prehaustorial resistance against wheat leaf rust in einkorn (Triticum monococcum), a progenitor of wheat. Front. Plant Sci. 2023, 14, 1252123. [Google Scholar] [CrossRef]
- Wu, H.; Kang, Z.; Li, X.; Li, Y.; Li, Y.; Wang, S.; Liu, D. Identification of Wheat Leaf Rust Resistance Genes in Chinese Wheat Cultivars and the Improved Germplasms. Plant Dis. 2020, 104, 2669–2680. [Google Scholar] [CrossRef]
- Hatta, M.A.M.; Steuernagel, B.; Wulff, B.B.H. Rapid gene cloning in wheat. In Applications of Genetic and Genomic Research in Cereals; Elsevier: Amsterdam, The Netherlands, 2019; pp. 65–95. [Google Scholar]
- Zhang, J.; Zhang, P.; Dodds, P.; Lagudah, E. How Target-Sequence Enrichment and Sequencing (TEnSeq) Pipelines Have Catalyzed Resistance Gene Cloning in the Wheat-Rust Pathosystem. Front. Plant Sci. 2020, 11, 678. [Google Scholar] [CrossRef]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef]
- Huang, L.; Brooks, S.A.; Li, W.; Fellers, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef]
- Lin, G.; Chen, H.; Tian, B.; Sehgal, S.K.; Singh, L.; Xie, J.; Rawat, N.; Juliana, P.; Singh, N.; Shrestha, S.; et al. Cloning of the broadly effective wheat leaf rust resistance gene Lr42 transferred from Aegilops tauschii. Nat. Commun. 2022, 13, 3044. [Google Scholar] [CrossRef]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef]
- Hewitt, T.; Zhang, J.; Huang, L.; Upadhyaya, N.; Li, J.; Park, R.; Hoxha, S.; McIntosh, R.; Lagudah, E.; Zhang, P. Wheat leaf rust resistance gene Lr13 is a specific Ne2 allele for hybrid necrosis. Mol. Plant 2021, 14, 1025–1028. [Google Scholar] [CrossRef]
- Yan, X.; Li, M.; Zhang, P.; Yin, G.; Zhang, H.; Gebrewahid, T.W.; Zhang, J.; Dong, L.; Liu, D.; Liu, Z.; et al. High-temperature wheat leaf rust resistance gene Lr13 exhibits pleiotropic effects on hybrid necrosis. Mol. Plant 2021, 14, 1029–1032. [Google Scholar] [CrossRef]
- Thind, A.K.; Wicker, T.; Šimková, H.; Fossati, D.; Moullet, O.; Brabant, C.; Vrána, J.; Doležel, J.; Krattinger, S.G. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat. Biotechnol. 2017, 35, 793–796. [Google Scholar] [CrossRef]
- Kolodziej, M.C.; Singla, J.; Sánchez-Martín, J.; Zbinden, H.; Šimková, H.; Karafiátová, M.; Doležel, J.; Gronnier, J.; Poretti, M.; Glauser, G.; et al. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nat. Commun. 2021, 12, 956. [Google Scholar] [CrossRef]
- Wang, Y.; Abrouk, M.; Gourdoupis, S.; Koo, D.-H.; Karafiátová, M.; Molnár, I.; Holušová, K.; Doležel, J.; Athiyannan, N.; Cavalet-Giorsa, E.; et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 2023, 55, 914–920. [Google Scholar] [CrossRef]
- Li, H.; Hua, L.; Zhao, S.; Hao, M.; Song, R.; Pang, S.; Liu, Y.; Chen, H.; Zhang, W.; Shen, T.; et al. Cloning of the broad-spectrum wheat leaf rust resistance gene Lr47 introgressed from Aegilops speltoides. Nat. Commun. 2023, 14, 6072. [Google Scholar] [CrossRef]
- Feuillet, C.; Schachermayr, G.; Keller, B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. For. Cell Mol. Biol. 1997, 11, 45–52. [Google Scholar] [CrossRef]
- O’Connell, R.J.; Panstruga, R. Tête à tête inside a plant cell: Establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 2006, 171, 699–718. [Google Scholar] [CrossRef]
- Hückelhoven, R. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef]
- Anker, C.C.; Niks, R.E. Prehaustorial resistance to the wheat leaf rust fungus, Puccinia triticina, in Triticum monococcum (s.s.). Euphytica 2001, 117, 209–215. [Google Scholar] [CrossRef]
- Niks, R.E.; Rubiales, D. Potentially durable resistance mechanisms in plants to specialised fungal pathogens. Euphytica 2002, 124, 201–216. [Google Scholar] [CrossRef]
- Skolotneva, E.; Salina, E. Resistance mechanisms involved in complex immunity of wheat against rust diseases. Vavilov J. Genet. Breed. 2019, 23, 542–550. [Google Scholar] [CrossRef]
- Rubiales, D.; Niks, R.E. Characterization of Lr34, a Major Gene Conferring Nonhypersensitive Resistance to Wheat Leaf Rust. Plant Dis. 1995, 79, 1208–1212. [Google Scholar] [CrossRef]
- Dmochowska-Boguta, M.; Alaba, S.; Yanushevska, Y.; Piechota, U.; Lasota, E.; Nadolska-Orczyk, A.; Karlowski, W.M.; Orczyk, W. Pathogen-regulated genes in wheat isogenic lines differing in resistance to brown rust Puccinia triticina. BMC Genom. 2015, 16, 742. [Google Scholar] [CrossRef]
- Orczyk, W.; Dmochowska-Boguta, M.; Czembor, H.J.; Nadolska-Orczyk, A. Spatiotemporal patterns of oxidative burst and micronecrosis in resistance of wheat to brown rust infection. Plant Pathol. 2010, 59, 567–575. [Google Scholar] [CrossRef]
- Serfling, A.; Templer, S.E.; Winter, P.; Ordon, F. Microscopic and Molecular Characterization of the Prehaustorial Resistance against Wheat Leaf Rust (Puccinia triticina) in Einkorn (Triticum monococcum). Front. Plant Sci. 2016, 7, 1668. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva, M.D.C.; Cardoso, F.; Planchon, S.; De Barros, D.; Renaut, J.; Ricardo, C. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6, 478. [Google Scholar] [CrossRef]
- Andrzejczak, O.A.; Sørensen, C.K.; Wang, W.Q.; Kovalchuk, S.; Hagensen, C.E.; Jensen, O.N.; Carciofi, M.; Hovmøller, M.S.; Rogowska-Wrzesinska, A.; Møller, I.M.; et al. The effect of phytoglobin overexpression on the plant proteome during nonhost response of barley (Hordeum vulgare) to wheat powdery mildew (Blumeria graminis f. sp. tritici). Sci. Rep. 2020, 10, 9192. [Google Scholar] [CrossRef]
- Wan, W.-L.; Kim, S.-T.; Castel, B.; Charoennit, N.; Chae, E. Genetics of autoimmunity in plants: An evolutionary genetics perspective. New Phytol. 2021, 229, 1215–1233. [Google Scholar] [CrossRef]
- Dyck, P.L.; Kerber, E.R. Resistance of the Race-Specific Type. In The Cereal Rusts, Volume II.; Diseases, Distribution, Epidemiology, and Control; Roelfs, A.P., Bushnell, W.R., Eds.; Academic Press: Orlando, FL, USA, 1985; pp. 469–500. [Google Scholar]
- Niks, R.E. Early abortion of colonies of leaf rust, Puccinia hordei, in partially resistant barley seedlings. Can. J. Bot. 1982, 60, 714–723. [Google Scholar] [CrossRef]
- Herrera-Foessel, S.A.; Lagudah, E.S.; Huerta-Espino, J.; Hayden, M.J.; Bariana, H.S.; Singh, D.; Singh, R.P. New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor. Appl. Genet. 2011, 122, 239–249. [Google Scholar] [CrossRef]
- Herrera-Foessel, S.A.; Singh, R.P.; Lillemo, M.; Huerta-Espino, J.; Bhavani, S.; Singh, S.; Lan, C.; Calvo-Salazar, V.; Lagudah, E.S. Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 2014, 127, 781–789. [Google Scholar] [CrossRef]
- Silva, M.C.; Nicole, M.; Guerra-GuimarÃes, L.; Rodrigues, C.J. Hypersensitive cell death and post-haustorial defence responses arrest the orange rust (Hemileia vastatrix) growth in resistant coffee leaves. Physiol. Mol. Plant Pathol. 2002, 60, 169–183. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Ryals, J.; Weymann, K.; Lawton, K.; Friedrich, L.; Ellis, D.; Steiner, H.Y.; Johnson, J.; Delaney, T.P.; Jesse, T.; Vos, P.; et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 1997, 9, 425–439. [Google Scholar] [CrossRef]
- Shah, J.; Tsui, F.; Klessig, D.F. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. MPMI 1997, 10, 69–78. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Manohar, M.; Tian, M.; Moreau, M.; Park, S.W.; Choi, H.W.; Fei, Z.; Friso, G.; Asif, M.; Manosalva, P.; von Dahl, C.C.; et al. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 2014, 5, 777. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of Salicylic Acid for the Induction of Systemic Acquired Resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic Acid, a Multifaceted Hormone to Combat Disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469.e16. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Holmes, E.C.; Rajniak, J.; Kim, J.G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4920–E4929. [Google Scholar] [CrossRef]
- Conrath, U. Systemic Acquired Resistance. Plant Signal. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, W.; Kinkema, M.; Li, X.; Dong, X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 1999, 96, 6523–6528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Trifa, Y.; Silva, H.; Pontier, D.; Lam, E.; Shah, J.; Klessig, D.F. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. MPMI 2000, 13, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Després, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Goritschnig, S.; Dong, X.; Li, X. A Gain-of-Function Mutation in a Plant Disease Resistance Gene Leads to Constitutive Activation of Downstream Signal Transduction Pathways in suppressor of npr1-1, constitutive 1. Plant Cell 2003, 15, 2636–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. For. Cell Mol. Biol. 2006, 48, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, T.; Sun, Y.; Zhang, Y.; Radojičić, A.; Ding, Y.; Tian, H.; Huang, X.; Lan, J.; Chen, S.; et al. Diverse Roles of the Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Plant Immunity. Plant Cell 2020, 32, 4002–4016. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-d.; Bi, W.-s.; Gao, J.; Yu, X.-m.; Wang, H.-y.; Liu, D.-q. Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agric. 2018, 17, 2468–2477. [Google Scholar] [CrossRef]
- Cantu, D.; Yang, B.; Ruan, R.; Li, K.; Menzo, V.; Fu, D.; Chern, M.; Ronald, P.C.; Dubcovsky, J. Comparative analysis of protein-protein interactions in the defense response of rice and wheat. BMC Genom. 2013, 14, 166. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Nyamesorto, B.; Luo, Y.; Mu, X.; Wang, F.; Kang, Z.; Lagudah, E.; Huang, L. A new mode of NPR1 action via an NB-ARC–NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 2020, 228, 959–972. [Google Scholar] [CrossRef]
- Prasad, P.; Savadi, S.; Bhardwaj, S.C.; Kashyap, P.L.; Gangwar, O.P.; Khan, H.; Kumar, S.; Kumar, R.; Patil, V. Stage-specific reprogramming of defense responsive genes during Lr24-mediated leaf rust resistance in wheat. J. Plant Pathol. 2019, 101, 283–293. [Google Scholar] [CrossRef]
- Doke, N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 1983, 23, 345–357. [Google Scholar] [CrossRef]
- Auh, C.K.; Murphy, T.M. Plasma Membrane Redox Enzyme Is Involved in the Synthesis of O2- and H2O2 by Phytophthora Elicitor-Stimulated Rose Cells. Plant Physiol. 1995, 107, 1241–1247. [Google Scholar] [CrossRef]
- Grant, M.; Brown, I.; Adams, S.; Knight, M.; Ainslie, A.; Mansfield, J. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. For. Cell Mol. Biol. 2000, 23, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Cheng, Z.; Ahmad, H.; Hayat, S. Reactive oxygen species (ROS) as defenses against a broad range of plant fungal infections and case study on ROS employed by crops against Verticillium dahliae wilts. J. Plant Interact. 2018, 13, 353–363. [Google Scholar] [CrossRef]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Montillet, J.L.; Chamnongpol, S.; Rustérucci, C.; Dat, J.; van de Cotte, B.; Agnel, J.P.; Battesti, C.; Inzé, D.; Van Breusegem, F.; Triantaphylidès, C. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005, 138, 1516–1526. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef]
- Nathan, C.; Cunningham-Bussel, A. Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 2013, 13, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lal, N.K.; Lin, Z.-J.D.; Ma, S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Nühse, T.S.; Bottrill, A.R.; Jones, A.M.; Peck, S.C. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. For. Cell Mol. Biol. 2007, 51, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef]
- Doke, N.; Miura, Y.; Sanchez, L.M.; Park, H.J.; Noritake, T.; Yoshioka, H.; Kawakita, K. The oxidative burst protects plants against pathogen attack: Mechanism and role as an emergency signal for plant bio-defence—A review. Gene 1996, 179, 45–51. [Google Scholar] [CrossRef]
- Mellersh, D.G.; Foulds, I.V.; Higgins, V.J.; Heath, M.C. H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. For. Cell Mol. Biol. 2002, 29, 257–268. [Google Scholar] [CrossRef] [PubMed]
- van Kan, J.A. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006, 11, 247–253. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Plotnikova, L.Y. The involvement of reactive oxygen species in defense of wheat lines with the genes introgressed from Agropyron species contributing the resistance against brown rust. Russ. J. Plant Physiol. 2009, 56, 181–189. [Google Scholar] [CrossRef]
- West, J.S.; Holdgate, S.; Townsend, J.A.; Edwards, S.G.; Jennings, P.; Fitt, B.D.L. Impacts of changing climate and agronomic factors on fusarium ear blight of wheat in the UK. Fungal Ecol. 2012, 5, 53–61. [Google Scholar] [CrossRef]
- Miedaner, T.; Juroszek, P. Climate change will influence disease resistance breeding in wheat in Northwestern Europe. Theor. Appl. Genet. 2021, 134, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising temperatures reduce global wheat production. Nat. Clim. Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Jevtić, R.; Župunski, V.; Lalošević, M.; Jocković, B.; Orbović, B.; Ilin, S. Diversity in susceptibility reactions of winter wheat genotypes to obligate pathogens under fluctuating climatic conditions. Sci. Rep. 2020, 10, 19608. [Google Scholar] [CrossRef]
- Albahri, G.; Alyamani, A.A.; Badran, A.; Hijazi, A.; Nasser, M.; Maresca, M.; Baydoun, E. Enhancing Essential Grains Yield for Sustainable Food Security and Bio-Safe Agriculture through Latest Innovative Approaches. Agronomy 2023, 13, 1709. [Google Scholar] [CrossRef]
- Naseri, B.; Sasani, S. Cultivar, planting date and weather linked to wheat leaf rust development. Cereal Res. Commun. 2020, 48, 203–210. [Google Scholar] [CrossRef]
- Naseri, B.; Jalilian, F. Characterization of leaf rust progress in wheat cultivars with different resistance levels and sowing dates. Eur. J. Plant Pathol. 2021, 159, 665–672. [Google Scholar] [CrossRef]
- Fenu, G.; Malloci, F.M. Forecasting Plant and Crop Disease: An Explorative Study on Current Algorithms. Big Data Cogn. Comput. 2021, 5, 2. [Google Scholar] [CrossRef]
- Chakraborty, S.; Murray, G.M.; Magarey, P.A.; Yonow, T.; O’Brien, R.G.; Croft, B.J.; Barbetti, M.J.; Sivasithamparam, K.; Old, K.M.; Dudzinski, M.J.; et al. Potential impact of climate change on plant diseases of economic significance to Australia. Australas. Plant Pathol. 1998, 27, 15–35. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012. [Google Scholar] [CrossRef]
- Sharma, P.K.; Ahmed, H.I.; Heuberger, M.; Koo, D.-H.; Quiroz-Chavez, J.; Adhikari, L.; Raupp, J.; Cauet, S.; Rodde, N.; Cravero, C.; et al. An online database for einkorn wheat to aid in gene discovery and functional genomics studies. Database 2023, 2023, baad079. [Google Scholar] [CrossRef] [PubMed]

| Effector Protein | Host | Localization | Function in Virulence | References |
|---|---|---|---|---|
| Pt2567 | Wheat | Secretion pathway | Inhibits programmed cell death and serves a non-toxic role in the infection of TcLr28. | [58] |
| Pt3 | Wheat | Unknown | Function in avirulence against wheat leaf rust in resistant genotypes. | [59] |
| Pt27 | Wheat | Unknown | Functions in avirulence against wheat leaf rust in resistant genotypes. | [59] |
| Pt18906 | Wheat | Nucleus and cytoplasm | Acts in the cytoplasm and may cause accumulation of reactive oxygen species and callose in TcLr10+27+31. | [55] |
| Pt13024 | Wheat | Nucleus and cytoplasm | Inhibits programmed cell death and triggers reactive oxygen species accumulation and callose deposition. | [56] |
| Pt_21 | Wheat | Apoplast | Suppresses host defense responses by directly targeting wheat TaTLP1 and inhibiting its anti-fungal activity. | [57] |
| Gene | Chromosome Position | R-Gene Product | R-Gene Class | Cloning Technique | References |
|---|---|---|---|---|---|
| Lr1 | 5DL | NLR | ASR | Map-based cloning | [104] |
| Lr9/Lr58 | 6BL/2BL | Tandem kinase–von Willebrand factor type-A domain fusion | ASR | MutIsoSeq | [114] |
| Lr10 | 1AS | NLR | ASR | Map-based cloning | [105] |
| Lr13/Ne2 | 2BS | NLR | APR | MutRenSeq | [110,111] |
| Lr14a | 7BL | Ankyrin transmembrane domain protein | ASR | MutChromSeq | [113] |
| Lr21 | 1DL | NLR | ASR | Map-based cloning | [106] |
| Lr22a | 2DS | NLR | APR | Map-based cloning and TACCA | [112] |
| Lr34/Yr18/Sr57 | 7DS | ATP-binding cassette transporter | APR | Map-based cloning | [107] |
| Lr42 | IDS | NLR | ASR | BSR-Seq mapping | [108] |
| Lr47 | 7AS | NLR | ASR | Map-based cloning EMTA approaches | [115] |
| Lr67/Yr46/Sr55 | 4DL | Anion transporter | APR | Map-based cloning | [109] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mapuranga, J.; Chang, J.; Zhao, J.; Liang, M.; Li, R.; Wu, Y.; Zhang, N.; Zhang, L.; Yang, W. The Underexplored Mechanisms of Wheat Resistance to Leaf Rust. Plants 2023, 12, 3996. https://doi.org/10.3390/plants12233996
Mapuranga J, Chang J, Zhao J, Liang M, Li R, Wu Y, Zhang N, Zhang L, Yang W. The Underexplored Mechanisms of Wheat Resistance to Leaf Rust. Plants. 2023; 12(23):3996. https://doi.org/10.3390/plants12233996
Chicago/Turabian StyleMapuranga, Johannes, Jiaying Chang, Jiaojie Zhao, Maili Liang, Ruolin Li, Yanhui Wu, Na Zhang, Lirong Zhang, and Wenxiang Yang. 2023. "The Underexplored Mechanisms of Wheat Resistance to Leaf Rust" Plants 12, no. 23: 3996. https://doi.org/10.3390/plants12233996
APA StyleMapuranga, J., Chang, J., Zhao, J., Liang, M., Li, R., Wu, Y., Zhang, N., Zhang, L., & Yang, W. (2023). The Underexplored Mechanisms of Wheat Resistance to Leaf Rust. Plants, 12(23), 3996. https://doi.org/10.3390/plants12233996






