Differential Responses of Medicago truncatula NLA Homologs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis
Abstract
:1. Introduction
2. Results
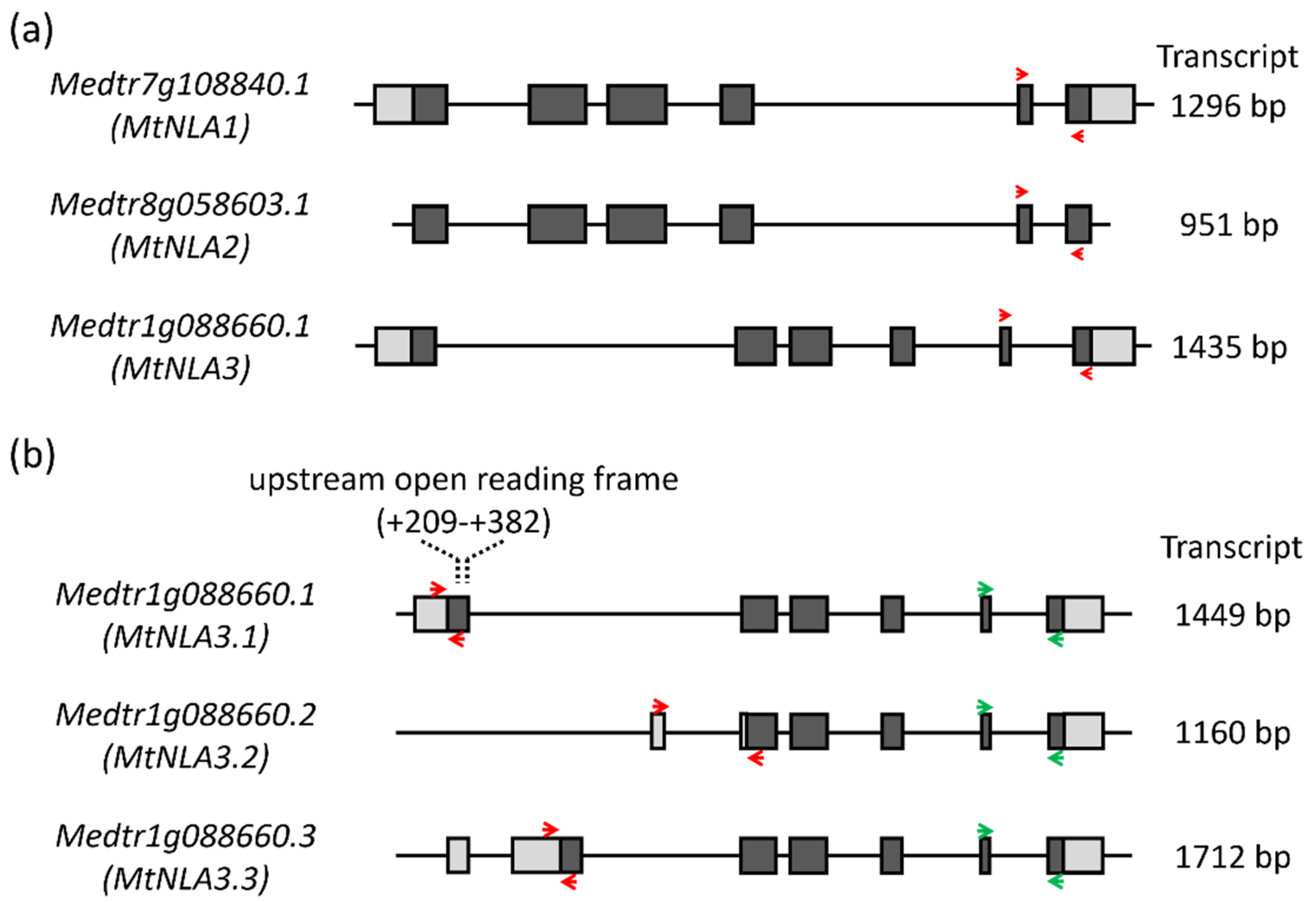
2.1. Comparison of the Sequence of MtNLA Paralogs and Variants
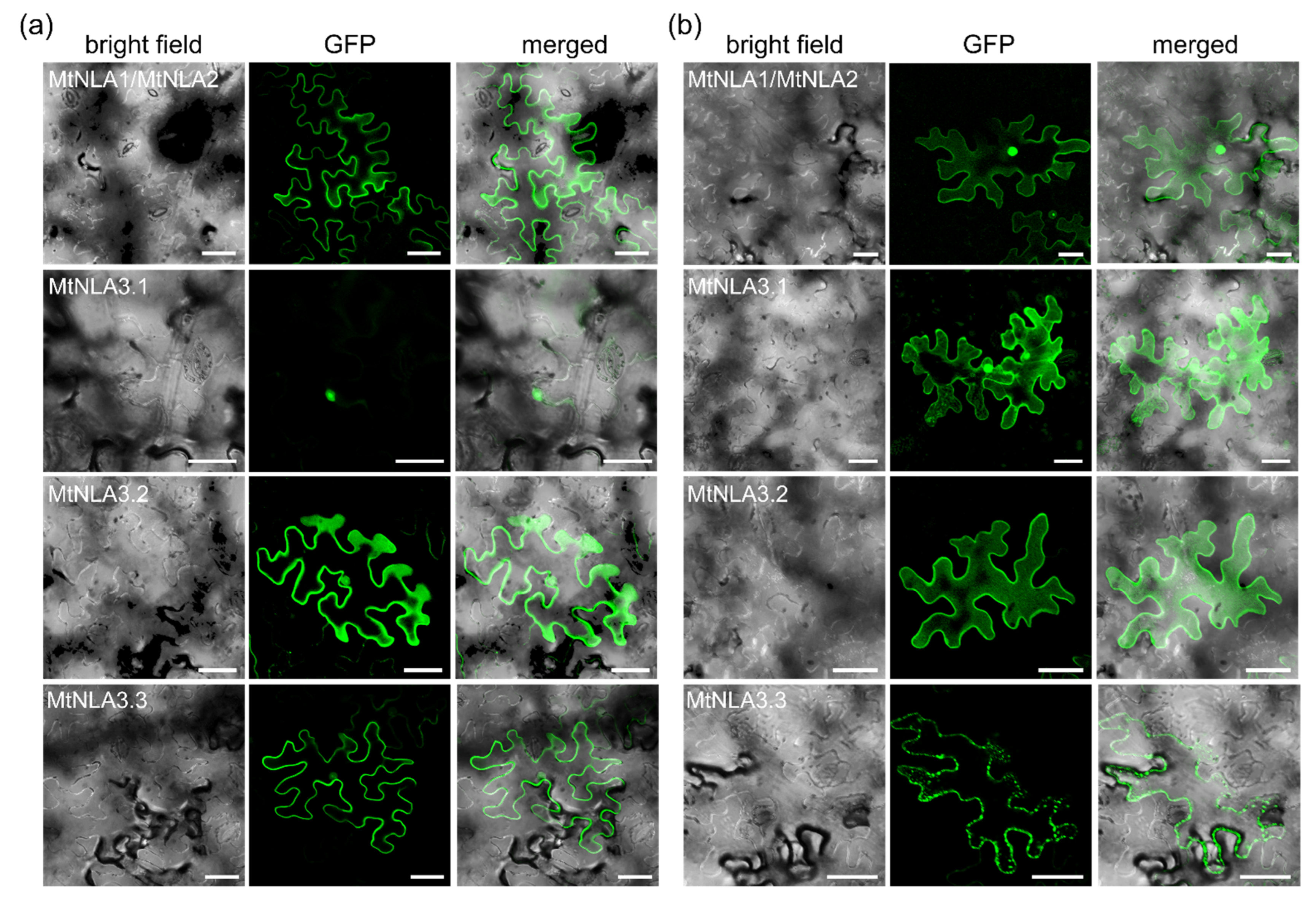
2.2. Subcellular Localization of MtNLA Paralogs and Variants
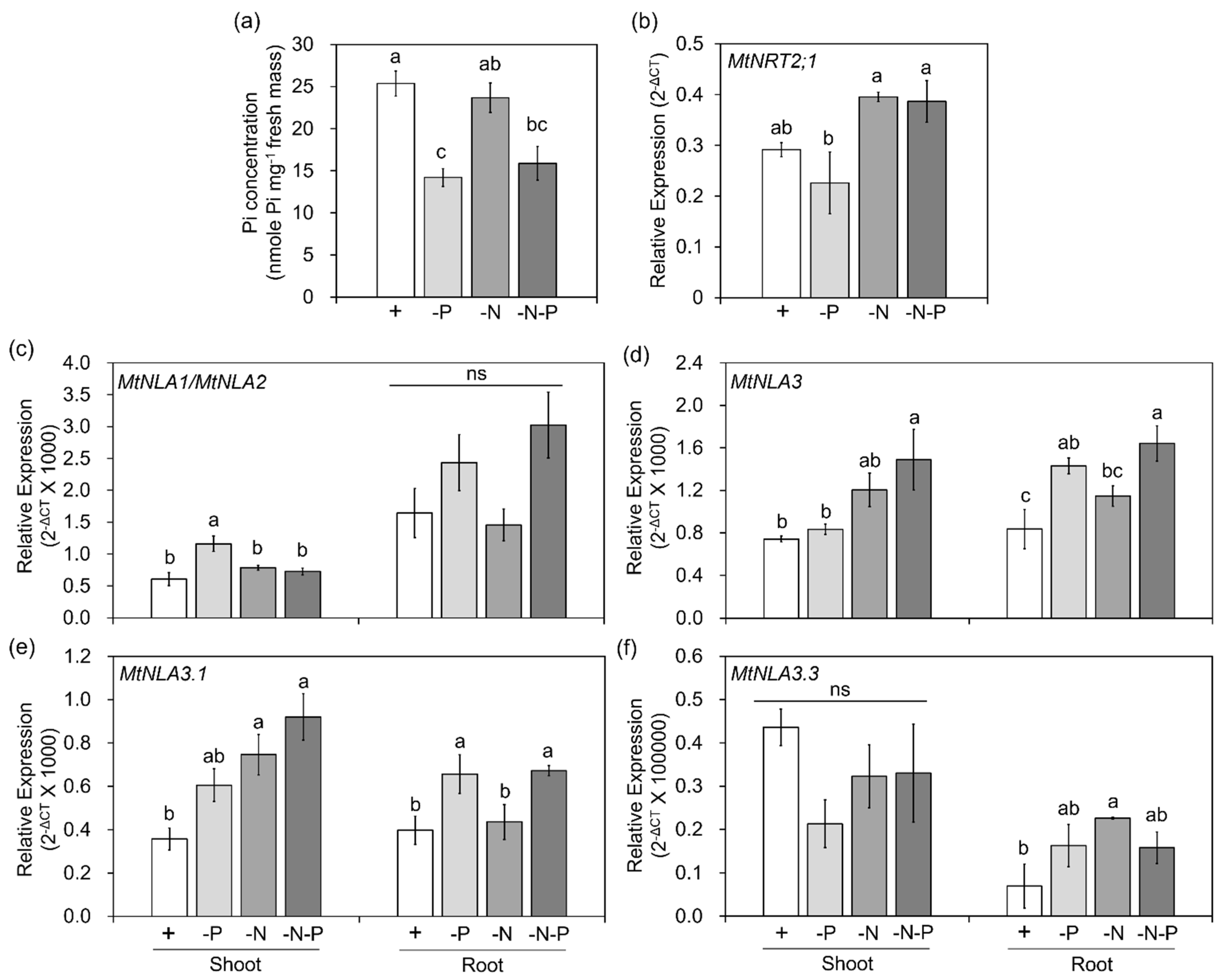
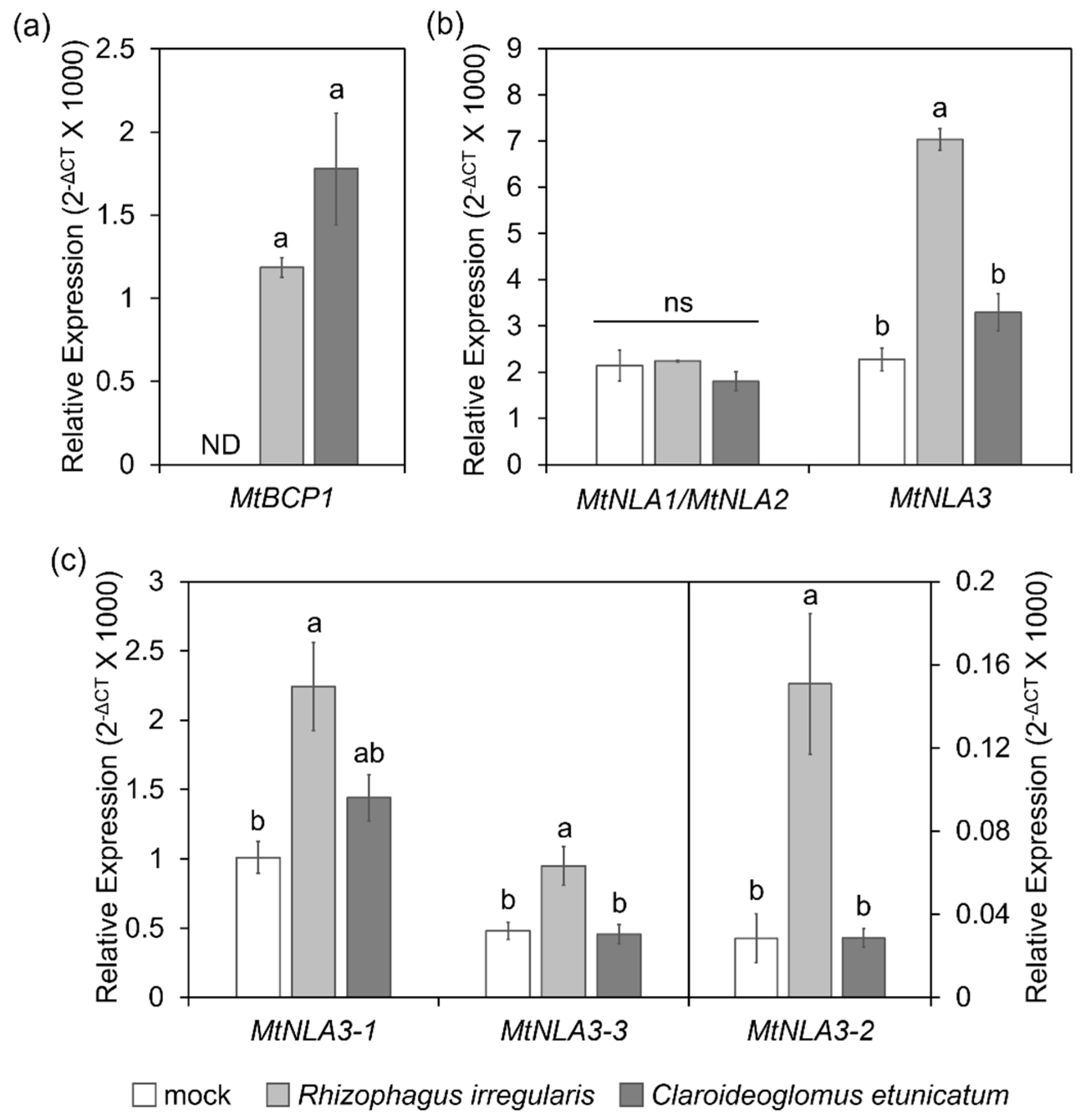
2.3. The Responses of MtNLA Paralogs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis
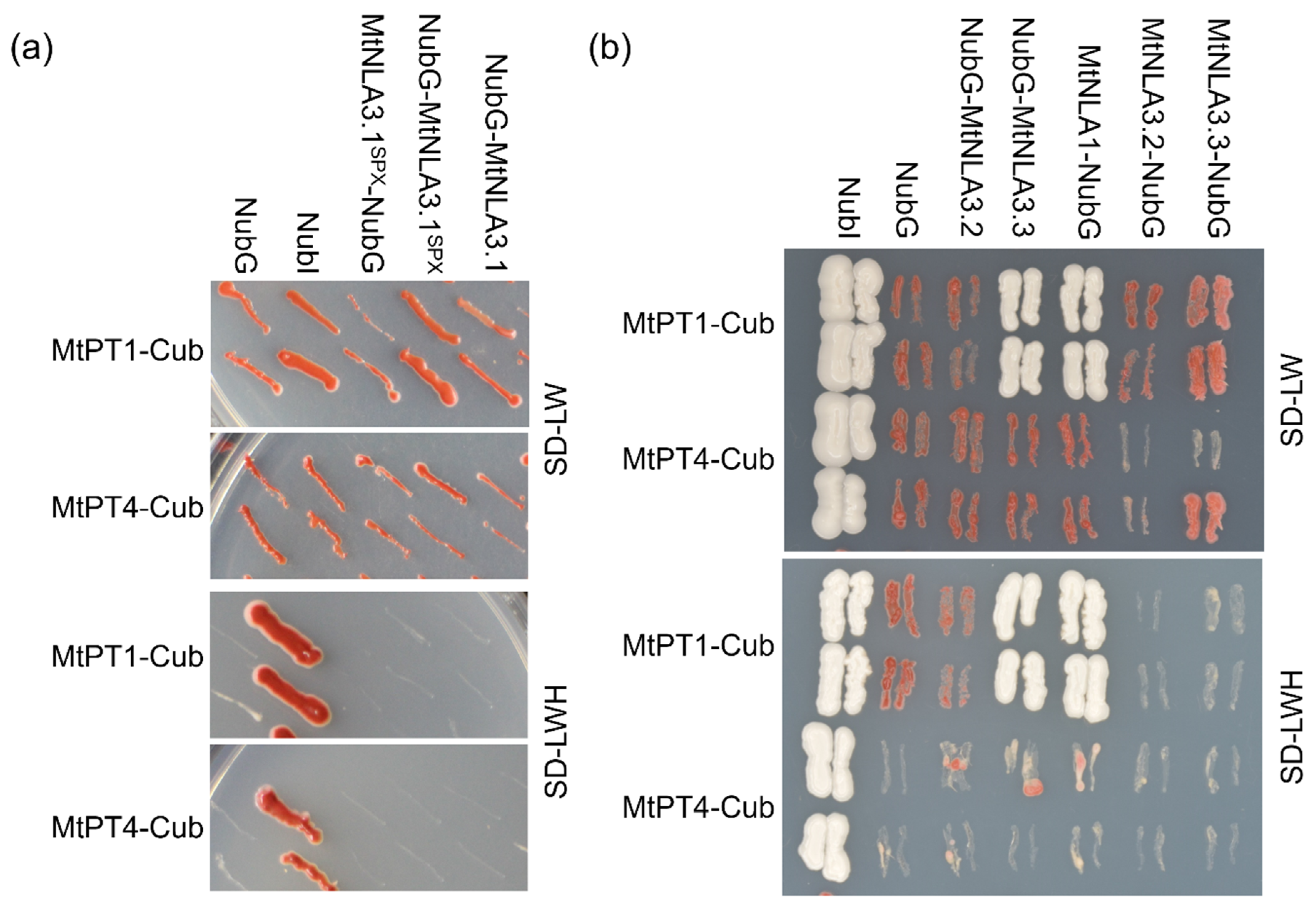
2.4. Interaction between MtNLAs and MtPTs
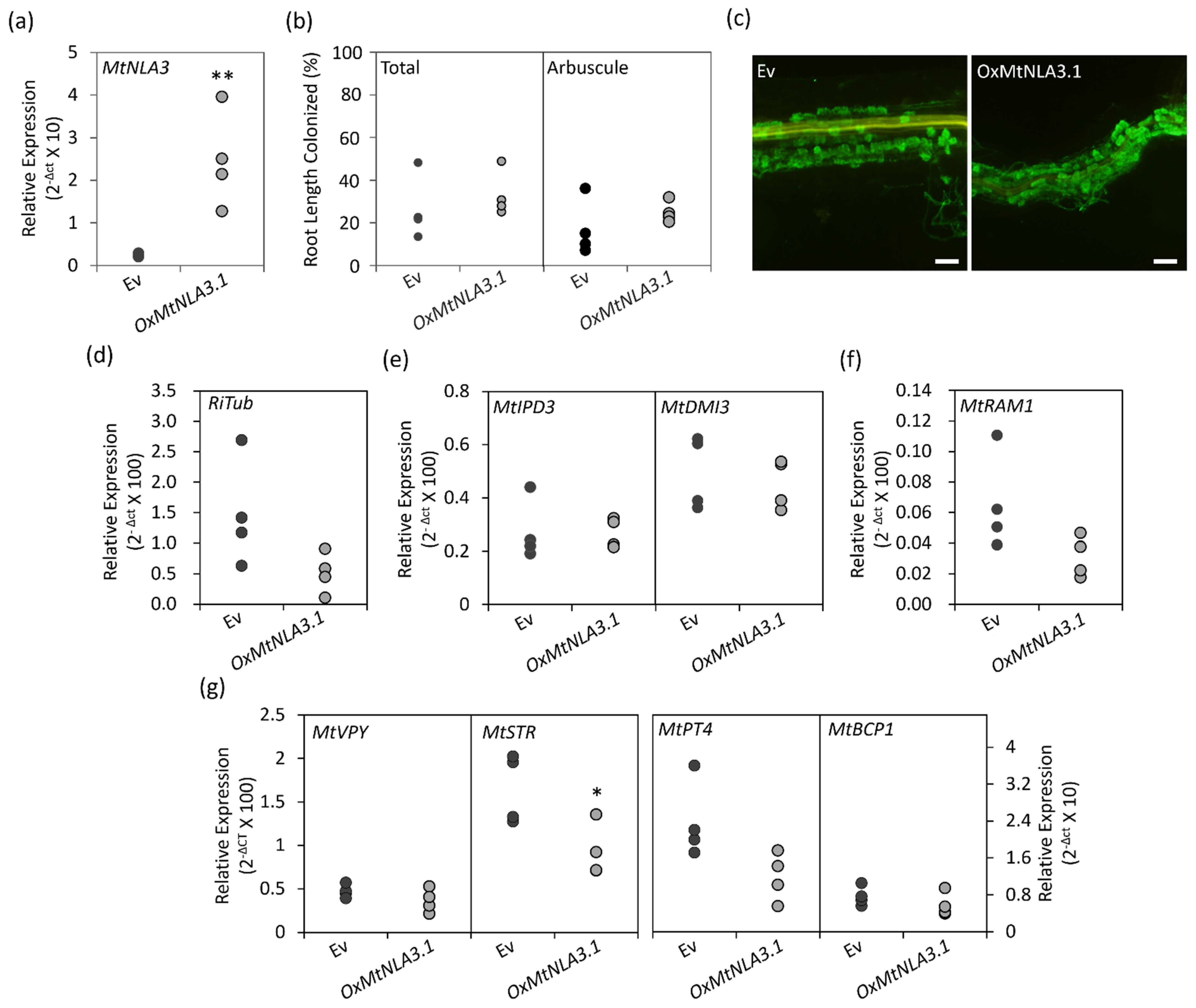
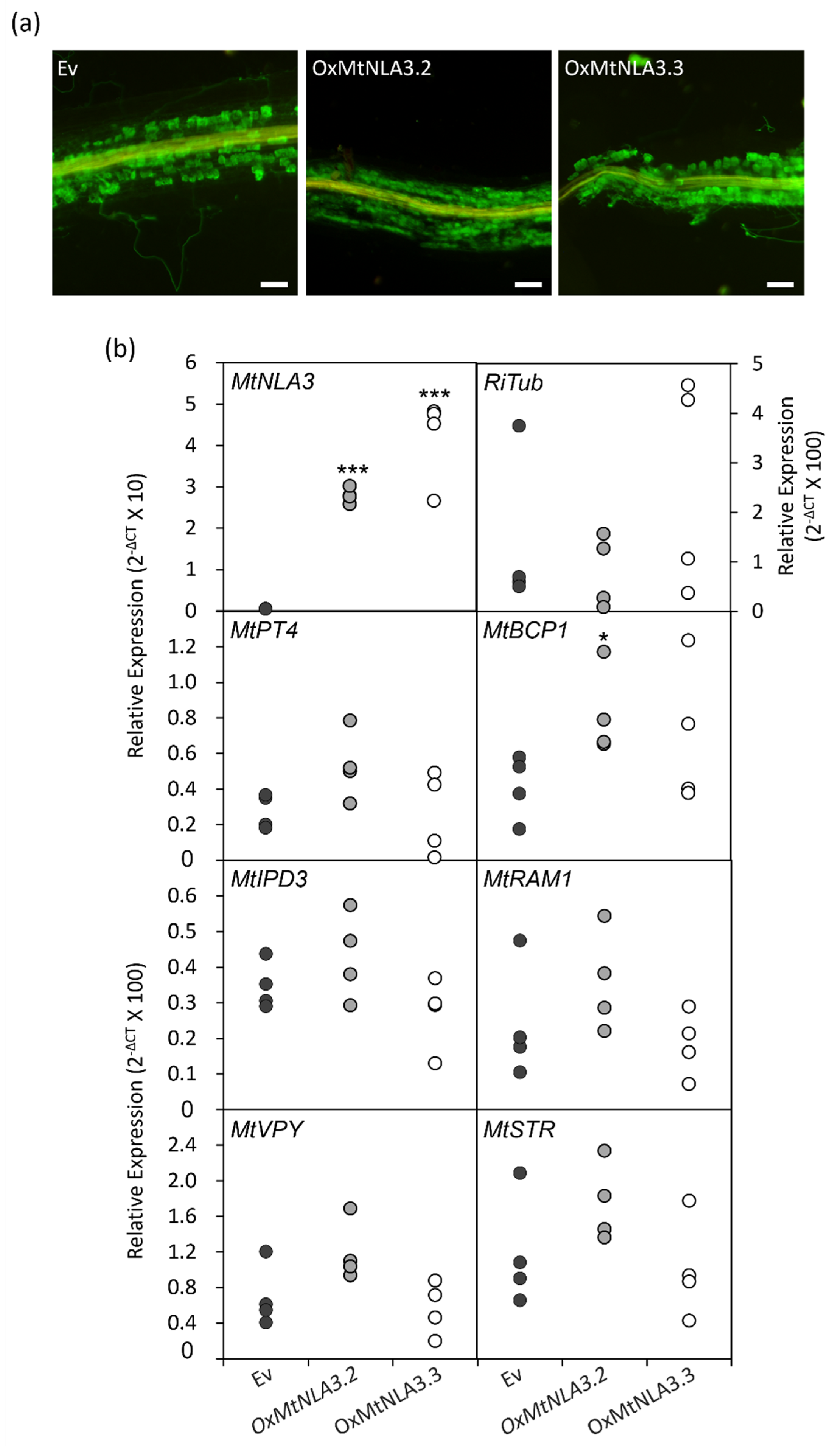
2.5. The Phenotype of MtNLA3-Overexpressing Roots in Response to Arbuscular Mycorrhizal Symbiosis
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Plasmid Construction
4.3. Agrobacterium Tumefaciens-Mediated Infiltration
4.4. Agrobacterium Rhizogenes-Mediated Root Transformation
4.5. Split-Ubiquitin Membrane Yeast Two-Hybrid Assay
4.6. WGA Staining and Analysis of Colonization Efficiency
4.7. Confocal Microscopy
4.8. RNA Isolation and Gene Expression Analysis
4.9. Pi Concentration Measurement
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, F.W.; Mudge, S.R.; Rae, A.L.; Glassop, D. Phosphate transport in plants. Plant Soil. 2003, 248, 71–83. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef]
- Chiou, T.-J.; Lin, S.-I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes. Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef]
- Schunmann, P.H.D.; Richardson, A.E.; Smith, F.W.; Delhaize, E. Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J. Exp. Bot. 2004, 55, 855–865. [Google Scholar] [CrossRef]
- Schunmann, P.H.D.; Richardson, A.E.; Vickers, C.E.; Delhaize, E. Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol. 2004, 136, 4205–4214. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 2005, 17, 3500–3512. [Google Scholar] [CrossRef]
- Bayle, V.; Arrighi, J.F.; Creff, A.; Nespoulous, C.; Vialaret, J.; Rossignol, M.; Gonzalez, E.; Paz-Ares, J.; Nussaume, L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 2011, 23, 1523–1535. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef]
- Huang, T.K.; Han, C.L.; Lin, S.I.; Chen, Y.J.; Tsai, Y.C.; Chen, Y.R.; Chen, J.W.; Lin, W.Y.; Chen, P.M.; Liu, T.Y.; et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane–localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.H. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.Y.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.X.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef]
- Hurlimann, H.C.; Pinson, B.; Stadler-Waibel, M.; Zeeman, S.C.; Freimoser, F.M. The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. Embo Rep. 2009, 10, 1003–1008. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y.; et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Guo, J.; Zhu, X.; Shi, J.; He, Q.; Liu, Y.; Wu, Y.; Zhang, L.; Lv, Q.; et al. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 2018, 219, 135–148. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef]
- Yue, W.; Ying, Y.; Wang, C.; Zhao, Y.; Dong, C.; Whelan, J.; Shou, H. OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 2017, 90, 1040–1051. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodrı’guez, J.; et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef]
- Wang, P.; Snijders, R.; Kohlen, W.; Liu, J.; Bisseling, T.; Limpens, E. Medicago SPX1 and SPX3 regulate phosphate homeostasis, mycorrhizal colonization, and arbuscule degradation. Plant Cell 2021, 33, 3470–3486. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, B.; Zheng, S.; Zhang, X.; Wang, X.; Dong, W.; Xie, Q.; Wang, G.; Xiao, Y.; Chen, F.; et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 2021, 184, 5527–5540. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.H.; Sun, C.; Liang, H.Y.; Wang, Y.; Bian, X.X.; Dong, C.Q.; Niu, X.F.; Yang, M.N.; Xu, G.H.; Chen, A.Q.; et al. SlSPX1-SlPHR complexes mediate the suppression of arbuscular mycorrhizal symbiosis by phosphate repletion in tomato. Plant Cell 2022, 34, 4045–4065. [Google Scholar] [CrossRef]
- Xie, X.; Lai, W.; Che, X.; Wang, S.; Ren, Y.; Hu, W.; Chen, H.; Tang, M. A SPX domain-containing phosphate transporter from Rhizophagus irregularis handles phosphate homeostasis at symbiotic interface of arbuscular mycorrhizas. New Phytol. 2022, 234, 650–671. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Hannam, C.; Gu, H.; Bi, Y.M.; Rothstein, S.J. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007, 50, 320–337. [Google Scholar] [CrossRef] [PubMed]
- Hannam, C.; Gidda, S.K.; Humbert, S.; Peng, M.; Cui, Y.; Dyer, J.M.; Rothstein, S.J.; Mullen, R.T. Distinct domains within the NITROGEN LIMITATION ADAPTATION protein mediate its subcellular localization and function in the nitrate-dependent phosphate homeostasis pathway. Botany 2018, 96, 79–96. [Google Scholar] [CrossRef]
- Liu, J.; Versaw, W.K.; Pumplin, N.; Gomez, S.K.; Blaylock, L.A.; Harrison, M.J. Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 2008, 283, 24673–24681. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.M.; Liu, J.L.; Li, Y.Y.; Zhang, J.; Li, S.X.; An, Y.R.; Hu, T.M.; Yang, P.Z. Functional analysis of the phosphate transporter gene MtPT6 from Medicago truncatula. Front. Plant Sci. 2021, 11, 620377. [Google Scholar] [CrossRef]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Zhang, X.; Noar, R.D.; Harrison, M.J. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. USA 2012, 109, E665–E672. [Google Scholar] [CrossRef]
- Volpe, V.; Giovannetti, M.; Sun, X.G.; Fiorilli, V.; Bonfante, P. The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non mycorrhizal roots. Plant Cell Environ. 2016, 39, 660–671. [Google Scholar] [CrossRef]
- Lin, W.Y.; Lin, Y.Y.; Chiang, S.F.; Syu, C.; Hsieh, L.C.; Chiou, T.J. Evolution of microRNA827 targeting in the plant kingdom. New Phytol. 2018, 217, 1712–1725. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Huang, T.K.; Tseng, C.Y.; Lai, Y.S.; Lin, S.I.; Lin, W.Y.; Chen, J.W.; Chiou, T.J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168–2183. [Google Scholar] [CrossRef]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Mao, C.; Lin, H. Characterization of the rice NLA family reveals a key role for OsNLA1 in phosphate homeostasis. Rice 2017, 10, 52. [Google Scholar] [CrossRef]
- Yang, S.Y.; Lu, W.C.; Ko, S.S.; Sun, C.M.; Hung, J.C.; Chiou, T.J. Upstream open reading frame and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiol. 2020, 182, 393–407. [Google Scholar] [CrossRef]
- Rahmat, Z.; Sohail, M.N.; Perrine-Walker, F.; Kaiser, B.N. Balancing nitrate acquisition strategies in symbiotic legumes. Planta 2023, 258, 12. [Google Scholar] [CrossRef]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Perez-Perez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.C.; Song, L.; Zhang, Y.; Zheng, Z.; Liu, D. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 2016, 170, 499–514. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Benedito, V.A.; Wang, M.Y.; Murray, J.D.; Zhao, P.X.; Tang, Y.H.; Udvardi, M.K. The Medicago truncatula gene expression atlas web server. BMC Bioinform. 2009, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Li, C.J.; Hsieh, C.Y.; Liu, L.Y.D.; Chen, Y.A.; Lin, W.Y. MtNF-YC6 and MtNF-YC11 are involved in regulating the transcriptional program of arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2022, 13, 976280. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Harrison, M.J. Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 2009, 151, 809–819. [Google Scholar] [CrossRef]
- Horvath, B.; Yeun, L.H.; Domonkos, A.; Halasz, G.; Gobbato, E.; Ayaydin, F.; Miro, K.; Hirsch, S.; Sun, J.H.; Tadege, M.; et al. Medicago truncatula IPD3 Is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant-Microbe Interact. 2011, 24, 1345–1358. [Google Scholar] [CrossRef]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Bornemann, S.; Morris, R.J.; Oldroyd, G.E.D. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef]
- Pumplin, N.; Mondo, S.J.; Topp, S.; Starker, C.G.; Gantt, J.S.; Harrison, M.J. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010, 61, 482–494. [Google Scholar] [CrossRef]
- Murray, J.D.; Duvvuru Muni, R.; Torres-Jerez, I.; Tang, Y.; Allen, S.; Andriankaja, M.; Li, G.; Laxmi, A.; Cheng, X.; Wen, J.; et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011, 65, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Gobbato, E.; Marsh, J.F.; Vernie, T.; Wang, E.; Maillet, F.; Kim, J.; Miller, J.B.; Sun, J.; Bano, S.A.; Ratet, P.; et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012, 22, 2236–2241. [Google Scholar] [CrossRef]
- Park, H.J.; Floss, D.S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M.J. Hyphal branching during arbuscule development requires Reduced Arbuscular Mycorrhiza1. Plant Physiol. 2015, 169, 2774–2788. [Google Scholar] [CrossRef]
- Zhang, Q.; Blaylock, L.A.; Harrison, M.J. Two Medicago truncatula Half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 2010, 22, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W.S. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Soltys-Kalina, D.; Szajko, K.; Stefańczyk, E.; Smyda-Dajmund, P.; Śliwka, J.; Marczewski, W. eQTL mapping of the 12S globulin cruciferin gene PGCRURSE5 as a novel candidate associated with starch content in potato tubers. Sci. Rep. 2020, 10, 17168. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Valenzuela, T.; Muñoz-Espinoza, C.; Riveros, A.; Pedreschi, R.; Arús, P.; Campos-Vargas, R.; Meneses, C. Expression QTL (eQTLs) analyses reveal candidate genes associated with fruit flesh softening rate in peach [Prunus persica (L.) Batsch]. Front. Plant Sci. 2019, 10, 1581. [Google Scholar] [CrossRef]
- Grunwald, U.; Guo, W.; Fischer, K.; Isayenkov, S.; Ludwig-Muller, J.; Hause, B.; Yan, X.; Kuster, H.; Franken, P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 2009, 229, 1023–1034. [Google Scholar] [CrossRef]
- Mateus, I.D.; Masclaux, F.G.; Aletti, C.; Rojas, E.C.; Savary, R.; Dupuis, C.; Sanders, I.R. Dual RNA-seq reveals large-scale non-conserved genotype x genotype-specific genetic reprograming and molecular crosstalk in the mycorrhizal symbiosis. ISME J. 2019, 13, 1226–1238. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Emmett, B.D.; Levesque-Tremblay, V.; MacLean, A.M.; Sun, X.; Satterlee, J.W.; Fei, Z.; Harrison, M.J. Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 2019, 42, 1758–1774. [Google Scholar] [CrossRef]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef]
- Vernie, T.; Camut, S.; Camps, C.; Rembliere, C.; de Carvalho-Niebel, F.; Mbengue, M.; Timmers, T.; Gasciolli, V.; Thompson, R.; le Signor, C.; et al. PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through Its ubiquitination activity in Medicago truncatula. Plant Physiol. 2016, 170, 2312–2324. [Google Scholar] [CrossRef]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Ivanov, S.; Harrison, M.J. A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 2014, 80, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Chabaud, M.; Garcia, F.; Becard, G.; Rosenberg, C.; Barker, D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 2001, 14, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Mcgonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method that gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Messinese, E.; Mun, J.H.; Yeun, L.H.; Jayaraman, D.; Rougé, P.; Barre, A.; Lougnon, G.; Schornack, S.; Bono, J.J.; Cook, D.R.; et al. A novel nuclear protein interacts with the symbiotic DMI3 calcium-and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 2007, 20, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Schäfer, M.; Li, D.; Halitschke, R.; Dong, C.; McGale, E.; Paetz, C.; Song, Y.; Li, S.; Dong, J.; et al. Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. Elife 2018, 7, e37093. [Google Scholar] [CrossRef]
- Lindsay, P.L.; Williams, B.N.; MacLean, A.; Harrison, M.J. A phosphate-dependent requirement for transcription factors IPD3 and IPD3L during arbuscular mycorrhizal symbiosis in Medicago truncatula. Mol. Plant-Microbe Interact. 2019, 32, 1277–1290. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, W.-Y.; Yang, H.-N.; Hsieh, C.-Y.; Deng, C. Differential Responses of Medicago truncatula NLA Homologs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis. Plants 2023, 12, 4129. https://doi.org/10.3390/plants12244129
Lin W-Y, Yang H-N, Hsieh C-Y, Deng C. Differential Responses of Medicago truncatula NLA Homologs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis. Plants. 2023; 12(24):4129. https://doi.org/10.3390/plants12244129
Chicago/Turabian StyleLin, Wei-Yi, Hsin-Ni Yang, Chen-Yun Hsieh, and Chen Deng. 2023. "Differential Responses of Medicago truncatula NLA Homologs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis" Plants 12, no. 24: 4129. https://doi.org/10.3390/plants12244129
APA StyleLin, W.-Y., Yang, H.-N., Hsieh, C.-Y., & Deng, C. (2023). Differential Responses of Medicago truncatula NLA Homologs to Nutrient Deficiency and Arbuscular Mycorrhizal Symbiosis. Plants, 12(24), 4129. https://doi.org/10.3390/plants12244129





