Analysis of Genetic Diversity in Adzuki Beans (Vigna angularis): Insights into Environmental Adaptation and Early Breeding Strategies for Yield Improvement
Abstract
1. Introduction
2. Results
2.1. Genotyping by Sequencing
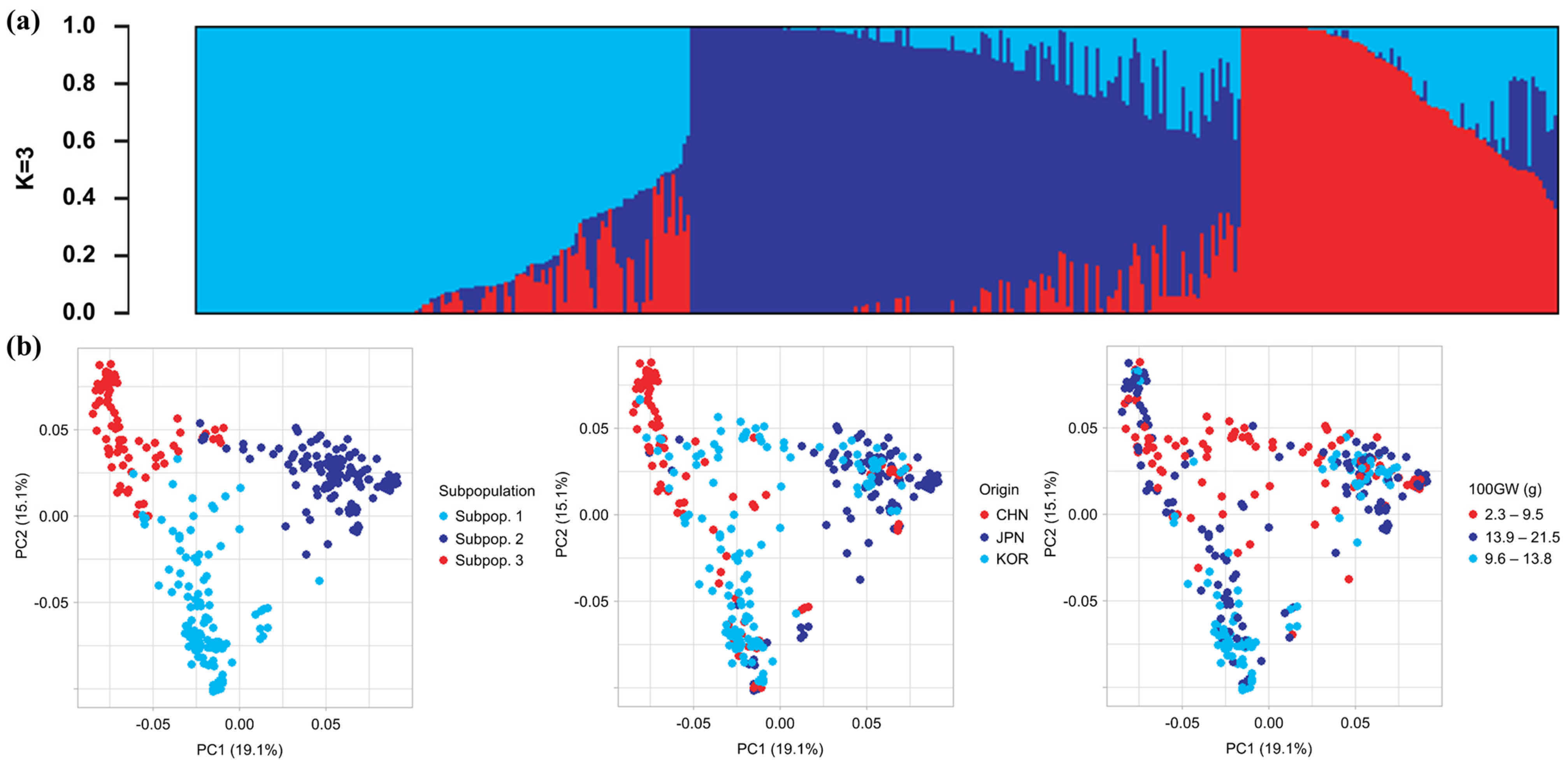
2.2. Population Structure
2.3. Genetic and Phenotypic Diversity
2.4. Gene Flow Analysis and Phylogenetic Analysis
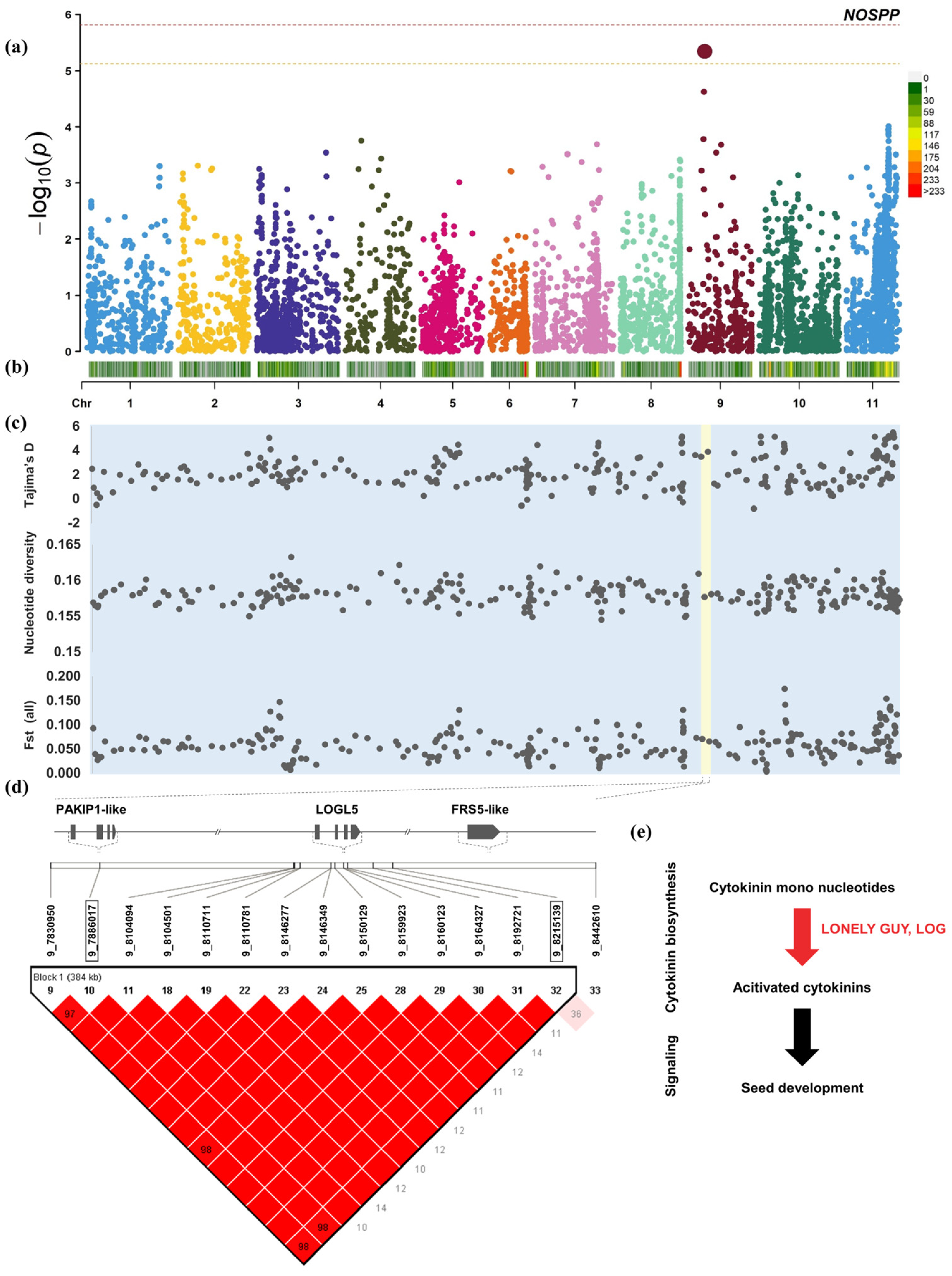
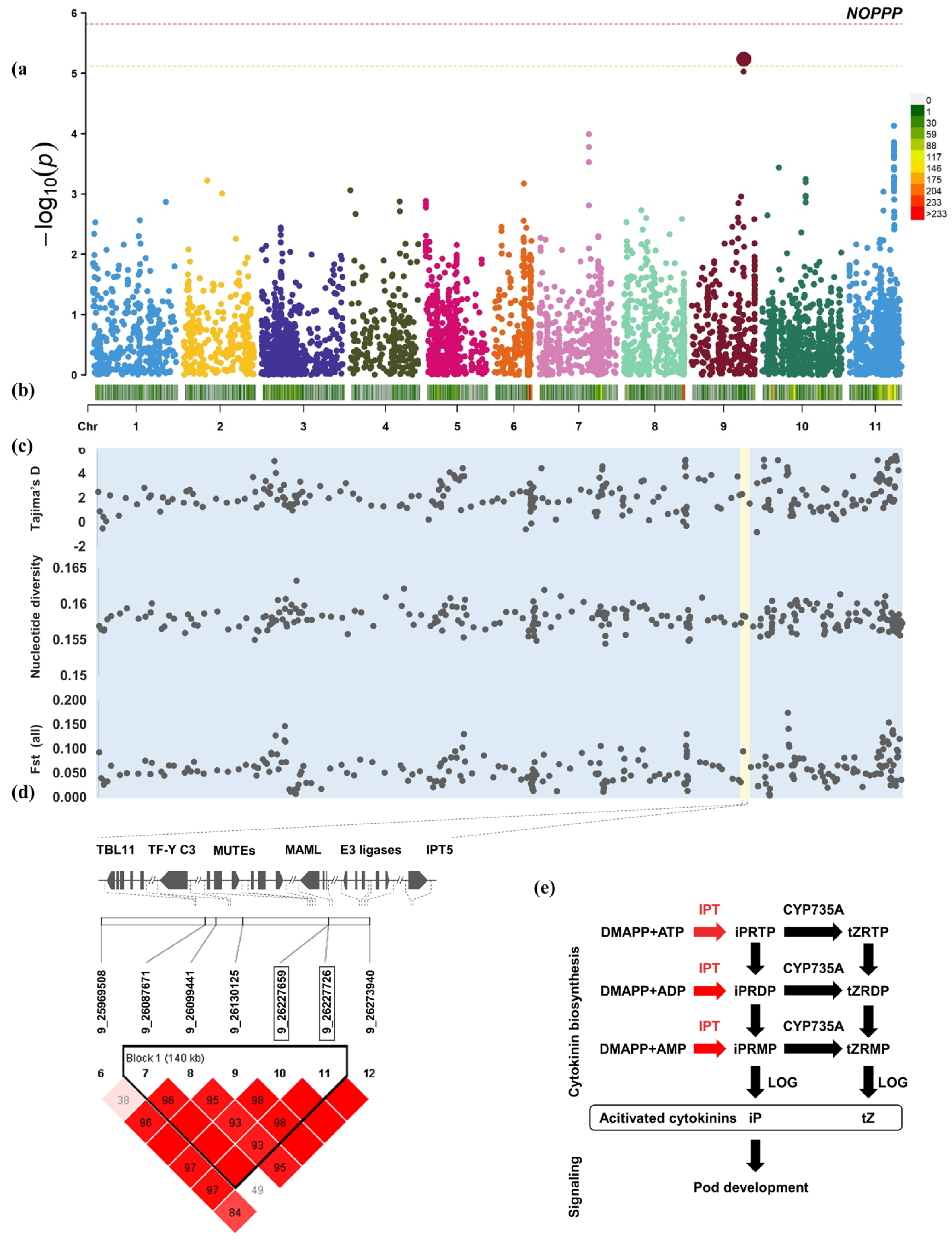
2.5. GWAS
3. Discussion
3.1. Migration History and Genetic Diversity Status of Adzuki Bean
3.2. Candidate Genes for Yield-Related Traits
3.3. Different Subpopulations Were Found to Have Different Traces of Domestication Selection on Yield-Related Traits in the Genome
4. Materials and Methods
4.1. Plant Material
4.2. DNA Extraction
4.3. Genotyping by Sequencing
4.4. Genome-Wide Association Study
4.5. Genetic Diversity and Differentiation Statistics
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, L.; Luo, G.; Zhu, X.; Wang, S.; Wang, L.; Cheng, X.; Chen, H. Genetic diversity and environmental influence on yield and yield-related traits of adzuki bean (Vigna angularis L.). Plants 2022, 11, 1132. [Google Scholar] [CrossRef] [PubMed]
- Crawford, G.; Underhill, A.; Zhao, Z.; Lee, G.; Feinman, G.; Nicholas, L.; Luan, F.; Yu, H.; Fang, H.; Cai, F. Late Neolithic plant remains from northern China: Preliminary results from Liangchengzhen, Shandong. Curr. Anthropol. 2005, 46, 309–317. [Google Scholar] [CrossRef]
- Liu, L.; Bestel, S.; Shi, J.; Song, Y.; Chen, X. Paleolithic human exploitation of plant foods during the last glacial maximum in North China. Proc. Natl. Acad. Sci. USA 2013, 110, 5380–5385. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, J.; Cheng, X. Adzuki Bean (Vigna angularis (Willd.) Ohwi & Ohashi) Breeding. In Advances in Plant Breeding Strategies: Legumes; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2019; pp. 1–23. [Google Scholar]
- Bi, S.; Wang, A.; Lao, F.; Shen, Q.; Liao, X.; Zhang, P.; Wu, J. Effects of frying, roasting and boiling on aroma profiles of adzuki beans (Vigna angularis) and potential of adzuki bean and millet flours to improve flavor and sensory characteristics of biscuits. Food Chem. 2021, 339, 127878. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.J.; Park, J.G.; Ahn, S.K.; Kim, K.W.; Choi, J.; Kim, H.Y.; Ha, S.-H.; Seo, W.D.; Kim, J.K. Discrimination of adzuki bean (Vigna angularis) geographical origin by targeted and non-targeted metabolite profiling with gas chromatography time-of-flight mass spectrometry. Metabolites 2020, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.J.; Kim, G.-H.; Lee, G.-A.; Lee, J.-R.; Cho, G.-T.; Ma, K.-H.; Lee, S. Antioxidant activities and total phenolic contents of three legumes. Korean J. Plant Resour. 2021, 34, 527–535. [Google Scholar]
- Lim, H.J.; Park, S.I.; Bak, S.G.; Cheong, S.H.; Lee, S.; Baek, Y.B.; Lee, C.M.; Lee, K.M.; Lee, S.W.; Lee, S.J. Beneficial effects of Vigna angularis extract in osteoporosis and osteoarthritis. Food Sci. Nutr. 2020, 8, 6550–6556. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bai, Z.; Wan, Y.; Shi, H.; Huang, X.; Nie, S. Antidiabetic effects of polysaccharide from azuki bean (Vigna angularis) in type 2 diabetic rats via insulin/PI3K/AKT signaling pathway. Food Hydrocoll. 2020, 101, 105456. [Google Scholar] [CrossRef]
- Yao, Y.; Xue, P.; Zhu, Y.; Gao, Y.; Ren, G. Antioxidant and immunoregulatory activity of polysaccharides from adzuki beans (Vigna angularis). Food Res. Int. 2015, 77, 251–256. [Google Scholar] [CrossRef]
- Kaga, A.; Isemura, T.; Tomooka, N.; Vaughan, D.A. The genetics of domestication of the azuki bean (Vigna angularis). Genetics 2008, 178, 1013–1036. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Bu, Y.; Wang, X.; Zhang, Y.; Guo, N.; Zhao, J.; Xing, H. Genome-wide association analysis for yield-related traits at the R6 stage in a Chinese soybean mini core collection. Genes Genom. 2021, 43, 897–912. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S. Traditional maintenance and multiplication of foxtail millet (Setaria italica (L.) P. Beauv.) landraces in China. Euphytica 1996, 87, 33–38. [Google Scholar] [CrossRef]
- Todd, S.E. The American Wheat Culturist: A Practical Treatise on the Culture of Wheat, Embracing a Brief History and Botanical Description of Wheat, with Full Practical Details for Selecting Seed, Producing New Varieties, and Cultivating on Different Kinds of Soil; Taintor Brothers & Company: New York, NY, USA, 1868. [Google Scholar]
- Alam, I.; Batool, K.; Huang, Y.; Liu, J.; Ge, L. Developing Genetic Engineering Techniques for Control of Seed Size and Yield. Int. J. Mol. Sci. 2022, 23, 13256. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Liu, X.; Pai, Q.; Wang, Y.; Wu, X. Molecular Network for Regulation of Seed Size in Plants. Int. J. Mol. Sci. 2023, 24, 10666. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Bai, Y.; Huang, W.; Wu, Y.; Yuan, Z.; Luan, X.; Liu, X.; Sun, L. Identification of potential gene regulatory pathways affecting the ratio of four-seed pod in soybean. Front. Genet. 2021, 12, 717770. [Google Scholar] [CrossRef] [PubMed]
- Araméndiz-Tatis, H.; Cardona-Ayala, C.; Espitia-Camacho, M. Heritability, genetic gain, and correlations in cowpea beans (Vigna unguiculata [L.] (Walp.). Rev. Colomb. Cienc. Hortícolas 2021, 15, e12321. [Google Scholar] [CrossRef]
- Bishop, J.; Garratt, M.; Breeze, T. Yield benefits of additional pollination to faba bean vary with cultivar, scale, yield parameter and experimental method. Sci. Rep. 2020, 10, 2102. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, M.; Shrestha, S.L.; Gautam, I.P.; Pandey, S. Evaluation of French Bean (Phaseolus vulgaris L.) varieties for summer season production in the mid–hills of central region of Nepal. Nepal. Hortic. 2020, 14, 48–55. [Google Scholar] [CrossRef]
- Vinay, H.; Jahnavi, A.; Rebasiddanavar, R.M.; Madhu, B. Influence of weather parameters on yield of Dolichos bean (Lablab purpureus L.). Pharma Innov. J. 2022, 11, 1556–1559. [Google Scholar]
- Palta, J.; Ludwig, C. Pod set and seed yield as affected by cytokinin application and terminal drought in narrow-leafed lupin. Aust. J. Agric. Res. 1997, 48, 81–90. [Google Scholar] [CrossRef]
- Yashima, Y.; Kaihatsu, A.; Nakajima, T.; Kokubun, M. Effects of source/sink ratio and cytokinin application on pod set in soybean. Plant Prod. Sci. 2005, 8, 139–144. [Google Scholar] [CrossRef]
- Chankaew, S.; Isemura, T.; Isobe, S.; Kaga, A.; Tomooka, N.; Somta, P.; Hirakawa, H.; Shirasawa, K.; Vaughan, D.A.; Srinives, P. Detection of genome donor species of neglected tetraploid crop Vigna reflexo pilosa (creole bean), and genetic structure of diploid species based on newly developed EST-SSR markers from azuki bean (Vigna angularis). PLoS ONE 2014, 9, e104990. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Gopalakrishna, T. Genetic diversity analysis in blackgram (Vigna mungo (L.) Hepper) using AFLP and transferable microsatellite markers from azuki bean (Vigna angularis (Willd.) Ohwi & Ohashi). Genome 2009, 52, 120–129. [Google Scholar] [PubMed]
- Isemura, T.; Noda, C.; Mori, S.; Yamashita, M.; Nakanishi, H.; Inoue, M.; Kamijima, O. Genetic variation and geographical distribution of Azuki bean (Vigna angularis) landraces based on the electrophoregram of seed storage proteins. Breed. Sci. 2001, 51, 225–230. [Google Scholar] [CrossRef][Green Version]
- Phansak, P.; Taylor, P.; Mongkolporn, O. Genetic diversity in yardlong bean (Vigna unguiculata ssp. sesquipedalis) and related Vigna species using sequence tagged microsatellite site analysis. Sci. Hortic. 2005, 106, 137–146. [Google Scholar]
- Xu, R.Q.; Tomooka, N.; Vaughan, D.A. AFLP markers for characterizing the azuki bean complex. Crop Sci. 2000, 40, 808–815. [Google Scholar] [CrossRef]
- Xu, H.; Jing, T.; Tomooka, N.; Kaga, A.; Isemura, T.; Vaughan, D. Genetic diversity of the azuki bean (Vigna angularis (Willd.) Ohwi & Ohashi) gene pool as assessed by SSR markers. Genome 2008, 51, 728–738. [Google Scholar]
- Danaisilichaichon, C.; Vejchasarn, P.; Patarapuwadol, S.; Tondelli, A.; Valè, G.; Toojinda, T.; Jantasuriyarat, C. Genome-Wide Association Study Using Genotyping by Sequencing for Bacterial Leaf Blight Resistance Loci in Local Thai Indica Rice. Agronomy 2023, 13, 1286. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, M.; Niu, X.; Wang, S.; Xu, Q.; Feng, Y.; Wang, C.; Deng, H.; Yuan, X.; Yu, H. Genetic variation and association mapping for 12 agronomic traits in indica rice. BMC Genom. 2015, 16, 1067. [Google Scholar] [CrossRef]
- Meng, B.; Wang, T.; Luo, Y.; Guo, Y.; Xu, D.; Liu, C.; Zou, J.; Li, L.; Diao, Y.; Gao, Z. Identification and allele combination analysis of rice grain shape-related genes by genome-wide association study. Int. J. Mol. Sci. 2022, 23, 1065. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Tayade, R.; Kang, B.-H.; Hahn, B.-S.; Ha, B.-K.; Kim, Y.-H. Genome-Wide Association Studies of Seven Root Traits in Soybean (Glycine max L.) Landraces. Int. J. Mol. Sci. 2023, 24, 873. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Tao, Y.; Xu, C.; Li, X.; Zhang, X.; Han, Y.; Yang, X.; Sun, J.; Li, W. Identification of genetic loci and candidate genes related to soybean flowering through genome wide association study. BMC Genom. 2019, 20, 987. [Google Scholar] [CrossRef] [PubMed]
- Noble, T.J.; Tao, Y.; Mace, E.S.; Williams, B.; Jordan, D.R.; Douglas, C.A.; Mundree, S.G. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front. Plant Sci. 2018, 8, 2102. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, Y.; Yang, Y.; Chi, Y.-J.; Zhou, J.; Chen, J.-Y.; Wang, F.; Fan, B.; Shi, K.; Zhou, Y.-H. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012, 159, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nie, X.; Tan, J.L.H.; Berger, F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15479–15484. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hua, L.; Dong, S.; Chen, H.; Zhu, X.; Jiang, J.e.; Zhang, F.; Li, Y.; Fang, X.; Chen, F. Os MAPK 6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 2015, 84, 672–681. [Google Scholar] [CrossRef]
- Liu, Z.; Mei, E.; Tian, X.; He, M.; Tang, J.; Xu, M.; Liu, J.; Song, L.; Li, X.; Wang, Z. OsMKKK70 regulates grain size and leaf angle in rice through the OsMKK4-OsMAPK6-OsWRKY53 signaling pathway. J. Integr. Plant Biol. 2021, 63, 2043–2057. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, W.; Shi, J.; Xu, J.; Zhang, D. MYB 56 encoding a R2 R 3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J. Integr. Plant Biol. 2013, 55, 1166–1178. [Google Scholar] [CrossRef]
- Yu, Y.-T.; Wu, Z.; Lu, K.; Bi, C.; Liang, S.; Wang, X.-F.; Zhang, D.-P. Overexpression of the MYB37 transcription factor enhances abscisic acid sensitivity, and improves both drought tolerance and seed productivity in Arabidopsis thaliana. Plant Mol. Biol. 2016, 90, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.; Liu, K.H.; Baldwin, G.S.; He, H. P21-activated kinase 1 stimulates colon cancer cell growth and migration/invasion via ERK-and AKT-dependent pathways. Biochim. Et Biophys. Acta BBA Mol. Cell Res. 2010, 1803, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, D.; Zhu, Y.; Li, W.; Chen, J.; Li, Y. Integrative transcriptomics and proteomics elucidate the regulatory mechanism of Hydrangea macrophylla flower-color changes induced by exogenous aluminum. Agronomy 2022, 12, 969. [Google Scholar] [CrossRef]
- Schwarz, I.; Scheirlinck, M.-T.; Otto, E.; Bartrina, I.; Schmidt, R.-C.; Schmülling, T. Cytokinin regulates the activity of the inflorescence meristem and components of seed yield in oilseed rape. J. Exp. Bot. 2020, 71, 7146–7159. [Google Scholar] [CrossRef] [PubMed]
- Zuñiga-Mayo, V.M.; Baños-Bayardo, C.R.; Díaz-Ramírez, D.; Marsch-Martínez, N.; de Folter, S. Conserved and novel responses to cytokinin treatments during flower and fruit development in Brassica napus and Arabidopsis thaliana. Sci. Rep. 2018, 8, 6836. [Google Scholar] [CrossRef]
- Chen, L.; Jameson, G.B.; Guo, Y.; Song, J.; Jameson, P.E. The LONELY GUY gene family: From mosses to wheat, the key to the formation of active cytokinins in plants. Plant Biotechnol. J. 2022, 20, 625–645. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Gao, Y.; Lu, G.; Habben, J.E.; Mao, G.; Chen, G.; Wang, J.; Yang, F.; Zhao, X. A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol. Biol. 2020, 102, 373–388. [Google Scholar] [CrossRef]
- Ma, L.; Li, G. FAR1-related sequence (FRS) and FRS-related factor (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef]
- Stirnberg, P.; Zhao, S.; Williamson, L.; Ward, S.; Leyser, O. FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 2012, 71, 907–920. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.-R. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, P.; Zhu, X.; Qi, K.; Xie, Z.; Zhang, H.; Li, X.; Gao, H.; Gu, T.; Gu, C. Acetylation of inorganic pyrophosphatase by S-RNase signaling induces pollen tube tip swelling by repressing pectin methylesterase. Plant Cell 2023, 35, 3544–3565. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Jikumaru, Y.; Hanada, A.; Nambara, E.; Abrams, S.R.; Kamiya, Y.; Seo, M. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shankar, R.; Yadav, S.K.; Kumar, V. Transcriptome analysis of ovules offers early developmental clues after fertilization in Cicer arietinum L. 3 Biotech 2023, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-K.; Qi, X.; Sugihara, K.; Dang, J.H.; Endo, T.A.; Miller, K.L.; Kim, E.-D.; Miura, T.; Torii, K.U. MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev. Cell 2018, 45, 303–315.e5. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.; Uosaki, H.; Shenje, L.T.; Kwon, C. Non-canonical Notch signaling: Emerging role and mechanism. Trends Cell Biol. 2012, 22, 257–265. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Li, Q.; Li, L.; Liu, Y.; Lv, Q.; Zhang, H.; Zhu, J.; Li, X. Influence of TaGW2-6A on seed development in wheat by negatively regulating gibberellin synthesis. Plant Sci. 2017, 263, 226–235. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Yang, Y.; Su, Q.; Li, Y.; Cheng, Z.; Song, Y.; Jin, X.; Wang, J. Fine mapping of a major QTL qHYF_B06 for peanut yield. Crop J. 2023, 11, 1533–1540. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef]
- Gui, J.; Liu, C.; Shen, J.; Li, L. Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 2014, 166, 1463–1478. [Google Scholar] [CrossRef]
- Gui, J.; Zheng, S.; Shen, J.; Li, L. Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 2015, 6, 804. [Google Scholar] [CrossRef] [PubMed]
- Baek, W.; Lim, C.W.; Lee, S.C. A DEAD-box RNA helicase, RH8, is critical for regulation of ABA signalling and the drought stress response via inhibition of PP2CA activity. Plant Cell Environ. 2018, 41, 1593–1604. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, T.; Huang, F.; Dai, P.; Cao, F.; Li, M. Ectopic expression of a R2R3 MYB transcription factor of dove tree (Davidia involucrata) aggravates seed abortion in Arabidopsis thaliana. Funct. Plant Biol. 2020, 47, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Z.; Ye, J.; Yang, Y.; Ye, J.; Xu, S.; Liu, J.; Yuan, X.; Wang, Y.; Zhang, M. Identification of SMG3, a QTL coordinately controls grain size, grain number per panicle, and grain weight in rice. Front. Plant Sci. 2022, 13, 880919. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liang, W.; Cui, X.; Chen, M.; Yin, C.; Luo, Z.; Zhu, J.; Lucas, W.J.; Wang, Z.; Zhang, D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J. 2015, 82, 570–581. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Dong, Y.; Yang, X.; Guo, D.; Qian, X.; Yan, F.; Wang, Y.; Li, J.; Wang, Q. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genome 2020, 63, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Knoth, C.; Eulgem, T. The oomycete response gene LURP1 is required for defense against Hyaloperonospora parasitica in Arabidopsis thaliana. Plant J. 2008, 55, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Otegui, M.S. ESCRT-mediated sorting and intralumenal vesicle concatenation in plants. Biochem. Soc. Trans. 2018, 46, 537–545. [Google Scholar] [CrossRef]
- Salem, M.A.; Li, Y.; Wiszniewski, A.; Giavalisco, P. Regulatory-associated protein of TOR (RAPTOR) alters the hormonal and metabolic composition of Arabidopsis seeds, controlling seed morphology, viability and germination potential. Plant J. 2017, 92, 525–545. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Lopes, K.V.; Apfata, J.A.; Fontes, E.P. NSP-interacting kinase, NIK: A transducer of plant defence signalling. J. Exp. Bot. 2010, 61, 3839–3845. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.D.; Reis, P.A.; Valente, M.A.S.; Irsigler, A.S.; Carvalho, C.M.; Loureiro, M.E.; Aragao, F.J.; Boston, R.S.; Fietto, L.G.; Fontes, E.P. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J. Biol. Chem. 2008, 283, 20209–20219. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.P.B. Insights into Regulatory Mechanisms of the NIK-Mediated Antiviral Defense: New Components and the Molecular Bases of the Defense. Ph.D. Thesis, Universidade Federal de Viçosa, Viçosa, Brazil, 2015. [Google Scholar]
- Malathi, V.; Renukadevi, P.; Rageshwari, S. Molecular Dynamics of Geminivirus-Host Interactome. In Plant Viruses: Diversity, Interaction and Managemen; CRC Press: Boca Raton, FL, USA, 2018; Chapter 10; pp. 173–194. [Google Scholar]
- Basu, D.; Tian, L.; Wang, W.; Bobbs, S.; Herock, H.; Travers, A.; Showalter, A.M. A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol. 2015, 15, 295. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.E.; Binkowski, K.A.; Olszewski, N.E. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 1996, 93, 9292–9296. [Google Scholar] [CrossRef] [PubMed]
- Rosado, A.; Schapire, A.L.; Bressan, R.A.; Harfouche, A.L.; Hasegawa, P.M.; Valpuesta, V.; Botella, M.A. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 2006, 142, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, E.S.; Chae, H.B.; Paeng, S.K.; Wi, S.D.; Bae, S.B.; Phan, K.A.T.; Lee, S.Y. Disulfide reductase activity of thioredoxin-h2 imparts cold tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2021, 568, 124–130. [Google Scholar] [CrossRef]
- Chandran, N.K.; Sriram, S.; Prakash, T. Identification of NBS-LRR resistance gene analogues (RGA) from rose (IIHRR13-4) resistant to powdery mildew (Podosphaera pannosa (Wallr.: Fr.) de Bary). J. Hortic. Sci. 2020, 15, 81–92. [Google Scholar] [CrossRef]
- Dang, P.M.; Lamb, M.C.; Chen, C.Y. Association of differentially expressed R-gene candidates with leaf spot resistance in peanut (Arachis hypogaea L.). Mol. Biol. Rep. 2021, 48, 323–334. [Google Scholar] [CrossRef]
- Nadeesha Lewke, B.; Komjanc, M.; Cestaro, A.; Cova, V.; Stefano, T.; Patocchi, A.; Michela, T.; Riccardo, V. Candidate Gene Identification for Rvi5 Apple Scab Resistance in Apple Cultivar ‘Murray’. In Proceedings of the International Symposium on Agriculture and Environment 2019, Wadduwa, Sri Lanka, 28 February 2019. [Google Scholar]
- Zhang, Y.; Zhang, Q.; Hao, L.; Wang, S.; Wang, S.; Zhang, W.; Xu, C.; Yu, Y.; Li, T. A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic. Res. 2019, 6, 93. [Google Scholar] [CrossRef]
- Shi, C.; Liu, H. How plants protect themselves from ultraviolet-B radiation stress. Plant Physiol. 2021, 187, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Rolly, N.K.; Yun, B.-W. Regulation of nitrate (NO3) transporters and glutamate synthase-encoding genes under drought stress in arabidopsis: The regulatory role of AtbZIP62 transcription factor. Plants 2021, 10, 2149. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, D.; Ma, L.; Jin, X.; Yang, P.; Ye, F.; Liu, P.; Gong, Z.; Wei, C. TMT-based quantitative proteomics analysis reveals the response of tea plant (Camellia sinensis) to fluoride. J. Proteom. 2018, 176, 71–81. [Google Scholar] [CrossRef]
- Bonza, C.; Carnelli, A.; De Michelis, M.I.; Rasi-Caldogno, F. Purification of the plasma membrane Ca2+-ATPase from radish seedlings by calmodulin-agarose affinity chromatography. Plant Physiol. 1998, 116, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Broedel, S.E., Jr.; Wolf, R.E., Jr. Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. J. Bacteriol. 1990, 172, 4023–4031. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. NADPH-generating dehydrogenases: Their role in the mechanism of protection against nitro-oxidative stress induced by adverse environmental conditions. Front. Environ. Sci. 2014, 2, 55. [Google Scholar] [CrossRef]
- Gupta, D.; Inouhe, M.; Rodríguez-Serrano, M.; Romero-Puertas, M.; Sandalio, L. Oxidative stress and arsenic toxicity: Role of NADPH oxidases. Chemosphere 2013, 90, 1987–1996. [Google Scholar] [CrossRef]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Gómez-Rodríguez, M.V.; Chaki, M.; Pedrajas, J.R.; Fernandez-Ocana, A.; Del Rio, L.A.; Barroso, J.B. The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant Cell Environ. 2006, 29, 1449–1459. [Google Scholar] [CrossRef]
- Dong, H.; Li, Y.; Fan, H.; Zhou, D.; Li, H. Quantitative proteomics analysis reveals resistance differences of banana cultivar ‘Brazilian’to Fusarium oxysporum f. sp. cubense races 1 and 4. J. Proteom. 2019, 203, 103376. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, L.; Gao, H.; Wenji, X.; Li, Q.; Li, H.; Gao, J. Comparative transcriptome analysis reveals heat stress-responsive genes and their signalling pathways in lilies (Lilium longiflorum vs. Lilium distichum). PLoS ONE 2020, 15, e0239605. [Google Scholar] [CrossRef] [PubMed]
- Dietzen, C.; Koprivova, A.; Whitcomb, S.J.; Langen, G.; Jobe, T.O.; Hoefgen, R.; Kopriva, S. The transcription factor EIL1 participates in the regulation of sulfur-deficiency response. Plant Physiol. 2020, 184, 2120–2136. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Ghantasala, S.; Roy Choudhury, S. Arabidopsis transmembrane receptor-like kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Liang, Y.; He, X.; Shen, Y.; Huang, Z. A novel ABA-responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in Arabidopsis thaliana. Plant Mol. Biol. Report. 2013, 31, 791–801. [Google Scholar] [CrossRef]
- Kurkela, S.; Borg-Franck, M. Structure and expression of kin2, one of two cold-and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 19, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Li, X.; Guo, M.; Deng, K.; Lin, J.; Tang, D.; Guo, X.; Liu, X. Regulation of salt and ABA responses by CIPK14, a calcium sensor interacting protein kinase in Arabidopsis. Sci. China Ser. C Life Sci. 2008, 51, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Que, Q.; Pan, R.; Wang, Q.; Gao, H.; Guan, X.; Che, J.; Lai, G. The single-stranded DNA-binding gene Whirly (Why1) with a strong pathogen-induced promoter from Vitis pseudoreticulata enhances resistance to Phytophthora capsici. Int. J. Mol. Sci. 2022, 23, 8052. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Wang, C.; Xiang, Y.; Qian, D. Current progress in plant V-ATPase: From biochemical properties to physiological functions. J. Plant Physiol. 2021, 266, 153525. [Google Scholar] [CrossRef]
- Feng, S.; Peng, Y.; Liu, E.; Ma, H.; Qiao, K.; Zhou, A.; Liu, S.; Bu, Y. Arabidopsis V-ATPase d2 subunit plays a role in plant responses to oxidative stress. Genes 2020, 11, 701. [Google Scholar] [CrossRef]
- Ji, M.G.; Park, H.J.; Cha, J.-Y.; Kim, J.A.; Shin, G.-I.; Jeong, S.Y.; Lee, E.S.; Yun, D.-J.; Lee, S.Y.; Kim, W.-Y. Expression of Arabidopsis thaliana Thioredoxin-h2 in Brassica napus enhances antioxidant defenses and improves salt tolerance. Plant Physiol. Biochem. 2020, 147, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Wei, N.; Hu, H.; Zhou, S.; Huang, Y.; Kong, Q.; Bie, Z.; Nie, W.-F.; Cheng, F. Thioredoxin h2 inhibits the MPKK5-MPK3 cascade to regulate the CBF–COR signaling pathway in Citrullus lanatus suffering chilling stress. Hortic. Res. 2023, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Su, P.-H.; Li, H.-m. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Tsuchiya, T.; Wang, X.J.; Beasley, B.; Cuzick, A.; Tör, M.; Zhu, T.; McDowell, J.M.; Holub, E.; Dangl, J.L. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007, 49, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ma, K.; Ji, G.; Pan, L.; Zhou, Q. Lipid transfer proteins involved in plant–pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 2022, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; An, J.; Xu, H.; Chen, J.; Pan, L.; Zhao, R.; Wang, N.; Gai, J.; Li, Y. A soybean sodium/hydrogen exchanger GmNHX6 confers plant alkaline salt tolerance by regulating Na+/K+ homeostasis. Front. Plant Sci. 2022, 13, 938635. [Google Scholar] [CrossRef]
- Sun, T.-J.; Fan, L.; Yang, J.; Cao, R.-Z.; Yang, C.-Y.; Zhang, J.; Wang, D.-M. A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na+ efflux rate and K+/Na+ ratio in Arabidopsis. BMC Plant Biol. 2019, 19, 469. [Google Scholar] [CrossRef]
- Al-Daoude, A.; de Torres Zabala, M.; Ko, J.-H.; Grant, M. RIN13 is a positive regulator of the plant disease resistance protein RPM1. Plant Cell 2005, 17, 1016–1028. [Google Scholar] [CrossRef]
- Du, D.; Zhang, C.; Xing, Y.; Lu, X.; Cai, L.; Yun, H.; Zhang, Q.; Zhang, Y.; Chen, X.; Liu, M. The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 2021, 19, 1052–1064. [Google Scholar] [CrossRef]
- Yan, W.; Jian, Y.; Duan, S.; Guo, X.; Hu, J.; Yang, X.; Li, G. Dissection of the Plant Hormone Signal Transduction Network in Late Blight-Resistant Potato Genotype SD20 and Prediction of Key Resistance Genes. Phytopathology 2023, 113, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; He, C.; Zhang, D.; Liu, X.; Xu, Z.; Tian, Y.; Liu, X.-H.; Zang, S.; Pauly, M.; Zhou, Y. Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 2017, 173, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, S.A.; Gille, S.; Pauly, M.; Krämer, U. Regulation of acetylation of plant cell wall components is complex and responds to external stimuli. Plant Signal. Behav. 2020, 15, 1687185. [Google Scholar] [CrossRef]
- Meng, H.; Sun, M.; Jiang, Z.; Liu, Y.; Sun, Y.; Liu, D.; Jiang, C.; Ren, M.; Yuan, G.; Yu, W. Comparative transcriptome analysis reveals resistant and susceptible genes in tobacco cultivars in response to infection by Phytophthora nicotianae. Sci. Rep. 2021, 11, 809. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Ma, Y.; Li, Y.; Li, X.; Liu, C.; Du, X.; Shi, G. Comparative transcriptome analysis revealed key factors for differential cadmium transport and retention in roots of two contrasting peanut cultivars. BMC Genom. 2018, 19, 938. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Osuna, D.; Colville, L.; Lorenzo, O.; Graeber, K.; Kuester, H.; Leubner-Metzger, G.; Kranner, I. Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS ONE 2013, 8, e78471. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.J.; Chintagunta, A.D.; Reddy, Y.M.; Rajjou, L.; Garlapati, V.K.; Agarwal, D.K.; Prasad, S.R.; Simal-Gandara, J. Implications of reactive oxygen and nitrogen species in seed physiology for sustainable crop productivity under changing climate conditions. Curr. Plant Biol. 2021, 26, 100197. [Google Scholar] [CrossRef]
- Gao, L.; Ma, Y.; Wang, P.; Wang, S.a.; Yang, R.; Wang, Q.; Li, L.; Li, Y. Transcriptome profiling of Clematis apiifolia: Insights into heat-stress responses. DNA Cell Biol. 2017, 36, 938–946. [Google Scholar] [CrossRef]
- Wei, S.; Song, Z.; Luo, S.; Zhong, Y.; Zhou, Y.; Lu, R. Transcriptome Analysis Reveals the Heat Stress Response Genes by Fire Stimulation in Michelia macclurei Dandy. Forests 2023, 14, 610. [Google Scholar] [CrossRef]
- Qian, W.; Xiao, B.; Wang, L.; Hao, X.; Yue, C.; Cao, H.; Wang, Y.; Li, N.; Yu, Y.; Zeng, J. CsINV5, a tea vacuolar invertase gene enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2018, 18, 228. [Google Scholar] [CrossRef]
- Szabados, L.; Kovács, H.; Zilberstein, A.; Bouchereau, A. Plants in extreme environments: Importance of protective compounds in stress tolerance. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2011; Volume 57, pp. 105–150. [Google Scholar]
- Wang, P.; Yang, Y.; Shi, H.; Wang, Y.; Ren, F. Small RNA and degradome deep sequencing reveal respective roles of cold-related microRNAs across Chinese wild grapevine and cultivated grapevine. BMC Genom. 2019, 20, 740. [Google Scholar] [CrossRef] [PubMed]
- Dyson, B.C.; Allwood, J.W.; Feil, R.; Xu, Y.; Miller, M.; Bowsher, C.G.; Goodacre, R.; Lunn, J.E.; Johnson, G.N. Acclimation of metabolism to light in A rabidopsis thaliana: The glucose 6-phosphate/phosphate translocator GPT 2 directs metabolic acclimation. Plant Cell Environ. 2015, 38, 1404–1417. [Google Scholar] [CrossRef] [PubMed]
- Weise, S.E.; Liu, T.; Childs, K.L.; Preiser, A.L.; Katulski, H.M.; Perrin-Porzondek, C.; Sharkey, T.D. Transcriptional regulation of the glucose-6-phosphate/phosphate translocator 2 is related to carbon exchange across the chloroplast envelope. Front. Plant Sci. 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Villarejo, A.; Srivastava, M.; Gelhaye, E.; Keech, O.; Droux, M.; Finkemeier, I.; Samuelsson, G.; Dietz, K.J.; Jacquot, J.-P. Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 2005, 7, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Klupczyńska, E.A.; Dietz, K.-J.; Małecka, A.; Ratajczak, E. Mitochondrial peroxiredoxin-IIF (PRXIIF) activity and function during seed aging. Antioxidants 2022, 11, 1226. [Google Scholar] [CrossRef] [PubMed]
- Zema, S.; Pelullo, M.; Nardozza, F.; Felli, M.P.; Screpanti, I.; Bellavia, D. A dynamic role of mastermind-like 1: A journey through the main (Path) ways between development and cancer. Front. Cell Dev. Biol. 2020, 8, 613557. [Google Scholar] [CrossRef] [PubMed]
- Manna, S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie 2015, 113, 93–99. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S.E.; Tang, Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559, 415–418. [Google Scholar] [CrossRef]
- Hirano, H. Basic 7S globulin in plants. J. Proteom. 2021, 240, 104209. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Reese, R.N.; Miller, M.A.; Rohila, J.S.; Subramanian, S.; Shen, Q.J.; Morandi, D.; Bücking, H.; Shulaev, V. A toolbox of genes, proteins, metabolites and promoters for improving drought tolerance in soybean includes the metabolite coumestrol and stomatal development genes. BMC Genom. 2016, 17, 102. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Tian, Z.; Chen, C.; Luo, L.; Zhao, B.; Wang, Z.; Yu, L.; Li, Y.; Sun, Y.; Li, W. Genome sequencing of adzuki bean (Vigna angularis) provides insight into high starch and low fat accumulation and domestication. Proc. Natl. Acad. Sci. USA 2015, 112, 13213–13218. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]
- Beerli, P.; Palczewski, M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics 2010, 185, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Aaron, M.; Charbon, G.; Lam, H.; Schwarz, H.; Vollmer, W.; Jacobs-Wagner, C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 2007, 64, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Pilhofer, M.; Rappl, K.; Eckl, C.; Bauer, A.P.; Ludwig, W.; Schleifer, K.-H.; Petroni, G. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J. Bacteriol. 2008, 190, 3192–3202. [Google Scholar] [CrossRef] [PubMed]
- Rane, C.K.; Minden, A. P21 activated kinases: Structure, regulation, and functions. Small GTPases 2014, 5, e28003. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Ishii, T.; Matsunaga, T.; Tominaga, R.; Kuromori, T.; Wada, T.; Shinozaki, K.; Hirayama, T. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 2008, 49, 1522–1535. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Cavell, A.C.; Dolan, L.; Roberts, K.; Grierson, C.S. Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 2000, 12, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Somerville, C.R. A dual mechanism of cellulose deficiency in shv3svl1. Plant Signal. Behav. 2016, 11, 110–124. [Google Scholar] [CrossRef]
- Yeats, T.H.; Sorek, H.; Wemmer, D.E.; Somerville, C.R. Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol. 2016, 171, 110–124. [Google Scholar] [CrossRef]
- Wu, B.; Meng, J.; Liu, H.; Mao, D.; Yin, H.; Zhang, Z.; Zhou, X.; Zhang, B.; Sherif, A.; Liu, H. Suppressing a phosphohydrolase of cytokinin nucleotide enhances grain yield in rice. Nat. Genet. 2023, 55, 1381–1389. [Google Scholar] [CrossRef]
- Kim, T.-W.; Guan, S.; Burlingame, A.L.; Wang, Z.-Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef]
- Saha, D.; Prasad, A.; Srinivasan, R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 2007, 45, 521–534. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177. [Google Scholar] [CrossRef]
- Chuang, H.-w.; Tseng, T.-S.; Hsieh, H.-Y.; Kao, T.-C.; Chen, G.-H. Common Cellular Events Implicated in the Regulation of Cold Stress Tolerance and Soft Rot Resistance Induced by Metabolites of Pseudomonas aeruginosa in Phalaenopsis orchids. Adv. Chem. Res. 2022, 1, 5–21. [Google Scholar] [CrossRef]
- Finkemeier, I.; Goodman, M.; Lamkemeyer, P.; Kandlbinder, A.; Sweetlove, L.J.; Dietz, K.-J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 2005, 280, 12168–12180. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Ren, T.; Niu, M.; Liu, X.; Liu, C.; Wang, H.; Yin, W.; Xia, X. Crucial Abiotic Stress Regulatory Network of NF-Y Transcription Factor in Plants. Int. J. Mol. Sci. 2023, 24, 4426. [Google Scholar] [CrossRef]
- Feng, J.; Liu, S.; Wang, M.; Lang, Q.; Jin, C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 2014, 240, 1335–1352. [Google Scholar] [CrossRef] [PubMed]

| Traits | Value | Subpopulation | Skewness | Kurtosis | ||
|---|---|---|---|---|---|---|
| 1 (KOR) | 2 (JPN) | 3 (CHN) | ||||
| Number of pods per plant | Region | 8.667–117.333 | 9.667–116 | 11.667–85.667 | 1.286 | 2.547 |
| Mean ± SD | 39.351 ± 17.184 ab | 36.194 ± 19.272 b | 42.345 ± 14.507 a | |||
| CV | 0.437 | 0.532 | 0.343 | |||
| Number of seeds per pod | Region | 4.4–9.5 | 4.4–9.9 | 4.6–9 | 0.140 | −0.365 |
| Mean ± SD | 6.678 ± 0.99 b | 7.178 ± 0.991 a | 6.889 ± 1.054 b | |||
| CV | 0.148 | 0.138 | 0.153 | |||
| 100-seed weight (g) | Region | 4.5–21.467 | 3.8–19.667 | 3.567–15.9 | −0.010 | −0.503 |
| Mean ± SD | 14.187 ± 3.192 a | 11.903 ± 3.685 b | 10.284 ± 2.576 c | |||
| CV | 0.225 | 0.310 | 0.250 | |||
| Yield per plant (g) | Region | 5.730–110.386 | 4.839–130.021 | 8.991–62.230 | 1.533 | 4.196 |
| Mean ± SD | 36.375 ± 16.134 a | 31.352 ± 21.522 b | 29.031 ± 11.192 b | |||
| CV | 0.444 | 0.686 | 0.386 | |||
| Flowering date (DAS) | Region | 47–77 | 44–77 | 47–76 | −0.214 | −0.778 |
| Mean ± SD | 63.466 ± 6.53 a | 61.054 ± 8.468 b | 60 ± 7.488 b | |||
| CV | 0.103 | 0.139 | 0.125 | |||
| Maturity date (DAS) | Region | 92–126 | 83–127 | 87–120 | −1.093 | 2.568 |
| Mean ± SD | 109.211 ± 4.998 a | 106.838 ± 6.546 b | 106.929 ± 6.446 b | |||
| CV | 0.046 | 0.061 | 0.060 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Choi, Y.-M.; Jeon, Y.-a.; Yi, J.; Shin, M.-J.; Desta, K.T.; Yoon, H. Analysis of Genetic Diversity in Adzuki Beans (Vigna angularis): Insights into Environmental Adaptation and Early Breeding Strategies for Yield Improvement. Plants 2023, 12, 4154. https://doi.org/10.3390/plants12244154
Wang X, Choi Y-M, Jeon Y-a, Yi J, Shin M-J, Desta KT, Yoon H. Analysis of Genetic Diversity in Adzuki Beans (Vigna angularis): Insights into Environmental Adaptation and Early Breeding Strategies for Yield Improvement. Plants. 2023; 12(24):4154. https://doi.org/10.3390/plants12244154
Chicago/Turabian StyleWang, Xiaohan, Yu-Mi Choi, Young-ah Jeon, JungYoon Yi, Myoung-Jae Shin, Kebede Taye Desta, and Hyemyeong Yoon. 2023. "Analysis of Genetic Diversity in Adzuki Beans (Vigna angularis): Insights into Environmental Adaptation and Early Breeding Strategies for Yield Improvement" Plants 12, no. 24: 4154. https://doi.org/10.3390/plants12244154
APA StyleWang, X., Choi, Y.-M., Jeon, Y.-a., Yi, J., Shin, M.-J., Desta, K. T., & Yoon, H. (2023). Analysis of Genetic Diversity in Adzuki Beans (Vigna angularis): Insights into Environmental Adaptation and Early Breeding Strategies for Yield Improvement. Plants, 12(24), 4154. https://doi.org/10.3390/plants12244154





