Characterization of Almond Scion/Rootstock Communication in Cultivar and Rootstock Tissues through an RNA-Seq Approach
Abstract
1. Introduction
2. Results
2.1. ‘Isabelona’ and ‘Lauranne’ Vigor Was Influenced by the Rootstock
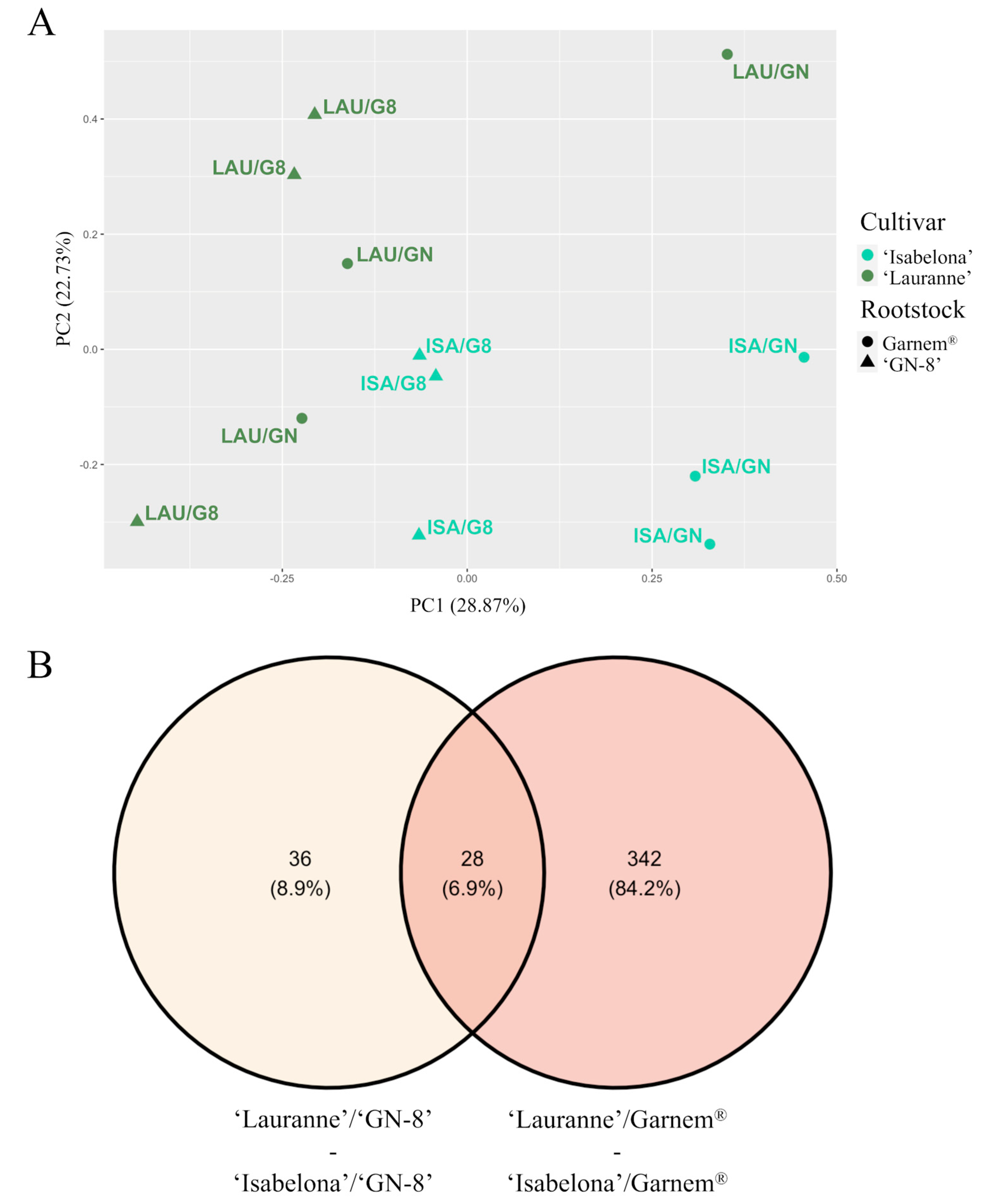
2.2. Rootstock Only Influenced Gene Expression in Combinations with ‘Isabelona’
2.3. Scion/Rootstock Interaction in Almond Affected Rootstock Molecular Profile
2.4. Garnem® Transcriptome Is More Affected by Cultivar Effect Than the ‘GN-8’ Transcriptome
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Phenotypic Data Collection in Nursery
4.3. RNA-Seq Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; von Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, Domestication, and Impacts on Shoot Phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef]
- Rubio-Cabetas, M.J.; Felipe, A.J.; Reighard, G.L. Rootstock Development. In Almonds: Botany, Production amd Uses; Socias i Company, R., Gradziel, T.M., Eds.; CABI: Wallingford, UK, 2017; pp. 193–220. [Google Scholar] [CrossRef]
- Aloni, B.; Cohen, R.; Karni, L.; Aktas, H.; Edelstein, M. Hormonal Signaling in Rootstock-Scion Interactions. Sci. Hortic. 2010, 127, 119–126. [Google Scholar] [CrossRef]
- Martínez-Ballesta, M.C.; Alcaraz-López, C.; Muries, B.; Mota-Cadenas, C.; Carvajal, M. Physiological Aspects of Rootstock-Scion Interactions. Sci. Hortic. 2010, 127, 112–118. [Google Scholar] [CrossRef]
- Albacete, A.; Martínez-Andújar, C.; Martínez-Pérez, A.; Thompson, A.J.; Dodd, I.C.; Pérez-Alfocea, F. Unravelling Rootstock×scion Interactions to Improve Food Security. J. Exp. Bot. 2015, 66, 2211–2226. [Google Scholar] [CrossRef]
- Foster, T.M.; Celton, J.M.; Chagne, D.; Tustin, D.S.; Gardiner, S.E. Two Quantitative Trait Loci, Dw1 and Dw2, Are Primarily Responsible for Rootstock-Induced Dwarfing in Apple. Hortic. Res. 2015, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Jiang, S.; Wang, F.; Tang, C.; Hao, N. An RNA-Seq Analysis of the Pear (Pyrus communis L.) Transcriptome, with a Focus on Genes Associated with Dwarf. Plant Gene 2015, 4, 69–77. [Google Scholar] [CrossRef][Green Version]
- López-Hinojosa, M.; de María, N.; Guevara, M.A.; Vélez, M.D.; Cabezas, J.A.; Díaz, L.M.; Mancha, J.A.; Pizarro, A.; Manjarrez, L.F.; Collada, C.; et al. Rootstock Effects on Scion Gene Expression in Maritime Pine. Sci. Rep. 2021, 11, 11582. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, C.W.; Gabel, A.; Hardcastle, T.J.; Robinson, S.; Miyashima, S.; Grosse, I.; Meyerowitz, E.M. Transcriptome Dynamics at Arabidopsis Graft Junctions Reveal an Intertissue Recognition Mechanism That Activates Vascular Regeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2447–E2456. [Google Scholar] [CrossRef] [PubMed]
- Wulf, K.E.; Reid, J.B.; Foo, E. Auxin Transport and Stem Vascular Reconnection—Has Our Thinking Become Canalized? Ann. Bot. 2019, 123, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, J.; Tan, M.; Mao, J.; An, N.; Sha, G.; Zhang, D.; Zhao, C.; Han, M. Transcriptome Analysis Reveals the Effects of Sugar Metabolism and Auxin and Cytokinin Signaling Pathways on Root Growth and Development of Grafted Apple. BMC Genom. 2016, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Umeda, M. Hormonal Control of Cell Division and Elongation along Differentiation Trajectories in Roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and Environmental Regulation of Root Development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An Auxin Gradient and Maximum in the Arabidopsis Root Apex Shown by High-Resolution Cell-Specific Analysis of IAA Distribution and Synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Friml, J. Auxin Regulates Distal Stem Cell Differentiation in Arabidopsis Roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef] [PubMed]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin Control of Root Development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001537. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H. Strigolactones Are Regulators of Root Development. New Phytol. 2011, 190, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tao, J.; Liu, S.; Huang, S.; Chen, S.; Xie, X.; Yoneyama, K.; Zhang, Y.; Xu, G. Strigolactones Are Involved in Phosphate- and Nitrate-Deficiency-Induced Root Development and Auxin Transport in Rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef]
- Jiang, L.; Matthys, C.; Marquez-Garcia, B.; De Cuyper, C.; Smet, L.; De Keyser, A.; Boyer, F.D.; Beeckman, T.; Depuydt, S.; Goormachtig, S. Strigolactones Spatially Influence Lateral Root Development through the Cytokinin Signaling Network. J. Exp. Bot. 2016, 67, 379–389. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A Master Regulator in Plant Root Development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef]
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496. [Google Scholar] [CrossRef]
- Jing, H.; Strader, L.C. Interplay of Auxin and Cytokinin in Lateral Root Development. Int. J. Mol. Sci. 2019, 20, 486. [Google Scholar] [CrossRef]
- Márquez, G.; Alarcón, M.V.; Salguero, J. Cytokinin Inhibits Lateral Root Development at the Earliest Stages of Lateral Root Primordium Initiation in Maize Primary Root. J. Plant Growth Regul. 2019, 38, 83–92. [Google Scholar] [CrossRef]
- Yaxley, J.R.; Ross, J.J.; Sherriff, L.J.; Reid, J.B. Gibberellin Biosynthesis Mutations and Root Development in Pea. Plant Physiol. 2001, 125, 627–633. [Google Scholar] [CrossRef]
- Ubeda-Tomás, S.; Swarup, R.; Coates, J.; Swarup, K.; Laplaze, L.; Beemster, G.T.S.; Hedden, P.; Bhalerao, R.; Bennett, M.J. Root Growth in Arabidopsis Requires Gibberellin/DELLA Signalling in the Endodermis. Nat. Cell Biol. 2008, 10, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Strauss, S.H.; Tsai, C.J.; Fang, K.; Chen, Y.; Jiang, X.; Busov, V.B. Gibberellins Regulate Lateral Root Formation in Populus through Interactions with Auxin and Other Hormones. Plant Cell 2010, 22, 623–639. [Google Scholar] [CrossRef]
- Wei, Z.; Li, J. Brassinosteroids Regulate Root Growth, Development, and Symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lei, W.; He, R.; Tang, X.; Han, J.; Zou, L.; Yin, Y.; Lin, H.; Zhang, D. Brassinosteroids Regulate Root Meristem Development by Mediating BIN2-UPB1 Module in Arabidopsis. PLoS Genet. 2020, 16, e1008883. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Negi, S.; Sukumar, P.; Muday, G.K. Ethylene Inhibits Lateral Root Development, Increases IAA Transport and Expression of PIN3 and PIN7 Auxin Efflux Carriers. Development 2011, 138, 3485–3495. [Google Scholar] [CrossRef]
- Qin, H.; He, L.; Huang, R. The Coordination of Ethylene and Other Hormones in Primary Root Development. Front. Plant Sci. 2019, 10, 874. [Google Scholar] [CrossRef]
- Farré, E.M. The Regulation of Plant Growth by the Circadian Clock. Plant Biol. 2012, 14, 401–410. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Circadian Clock during Plant Development. J. Plant Res. 2018, 131, 59–66. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome Regulation of Branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef]
- Rausenberger, J.; Tscheuschler, A.; Nordmeier, W.; Wüst, F.; Timmer, J.; Schäfer, E.; Fleck, C.; Hiltbrunner, A. Photoconversion and Nuclear Trafficking Cycles Determine Phytochrome A’s Response Profile to Far-Red Light. Cell 2011, 146, 813–825. [Google Scholar] [CrossRef]
- Casal, J.J. Shade Avoidance. Arab. B 2012, 10, e0157. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.K.; Finlayson, S.A. Phytochrome B Promotes Branching in Arabidopsis by Suppressing Auxin Signaling. Plant Physiol. 2014, 164, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Holalu, S.V.; Finlayson, S.A. The Ratio of Red Light to Far Red Light Alters Arabidopsis Axillary Bud Growth and Abscisic Acid Signalling before Stem Auxin Changes. J. Exp. Bot. 2017, 68, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, Á.; Thorp, G.; Grimplet, J.; Rubio-Cabetas, M. Phenotyping Almond Orchards for Architectural Traits Influenced by Rootstock Choice. Horticulturae 2021, 7, 159. [Google Scholar] [CrossRef]
- Montesinos, Á.; Grimplet, J.; Rubio-Cabetas, M.J. Proleptic and Sylleptic Shoot Formation Is Affected by Rootstock Genotype in Two-Year-Old Branches of Almond Trees. Agronomy 2022, 12, 2006. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual Regulation Role of GH3.5 in Salicylic Acid and Auxin Signaling during Arabidopsis-Pseudomonas Syringae Interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, D.; Song, Y.; Jin, C.; Ji, J.; Wang, G.; Li, X.; Guan, C. Enhancement of Methyl Salicylate Accumulation Promotes Early Flowering in Transgenic Tobacco Plants by Overexpressing a Carboxymethyl Transferase (SAMT) Gene from Lycium Chinense. Mol. Breed. 2020, 40, 52. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Köhler, C. Auxin Production Couples Endosperm Development to Fertilization. Nat. Plants 2015, 1, 15184. [Google Scholar] [CrossRef] [PubMed]
- Pierdonati, E.; Unterholzner, S.J.; Salvi, E.; Svolacchia, N.; Bertolotti, G.; Dello Ioio, R.; Sabatini, S.; Di Mambro, R. Cytokinin-Dependent Control of GH3 Group II Family Genes in the Arabidopsis Root. Plants 2019, 8, 94. [Google Scholar] [CrossRef]
- Barbier, F.F.; Lunn, J.E.; Beveridge, C.A. Ready, Steady, Go! A Sugar Hit Starts the Race to Shoot Branching. Curr. Opin. Plant Biol. 2015, 25, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.L.; Hollender, C.A. Branching out: New Insights into the Genetic Regulation of Shoot Architecture in Trees. Curr. Opin. Plant Biol. 2019, 47, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhuang, W.; Tu, X.; Gao, Z.; Xiong, A.; Yu, X.; Li, X.; Li, F.; Qu, S. Transcriptomic analysis of interstock-induced dwarfism in Sweet Persimmon (Diospyros kaki Thunb.). Hortic. Res. 2019, 6, 51. [Google Scholar] [CrossRef]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.P. Distinct and Overlapping Roles of Two Gibberellin 3-Oxidases in Arabidopsis Development. Plant J. 2006, 45, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Joseph, M.P.; Papdi, C.; Kozma-Bognár, L.; Nagy, I.; López-Carbonell, M.; Rigó, G.; Koncz, C.; Szabados, L. The Arabidopsis ZINC FINGER PROTEIN3 Interferes with Abscisic Acid and Light Signaling in Seed Germination and Plant Development. Plant Physiol. 2014, 165, 1203–1220. [Google Scholar] [CrossRef]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.C.; Kim, H.S.; Kim, S. MYB3 Plays an Important Role in Lignin and Anthocyanin Biosynthesis under Salt Stress Condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, B.; Li, A.; Chu, C. NRT1.1s in Plants: Functions beyond Nitrate Transport. J. Exp. Bot. 2020, 71, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Avramova, Z.; Fromm, M. The Arabidopsis Trithorax-like Factor ATX1 Functions in Dehydration Stress Responses via ABA-Dependent and ABA-Independent Pathways. Plant J. 2011, 66, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Jang, Y.J.; Park, O.K. AP2/ERF Family Transcription Factors ORA59 and RAP2.3 Interact in the Nucleus and Function Together in Ethylene Responses. Front. Plant Sci. 2018, 9, 1675. [Google Scholar] [CrossRef]
- León, J.; Costa-Broseta, Á.; Castillo, M.C. RAP2.3 Negatively Regulates Nitric Oxide Biosynthesis and Related Responses through a Rheostat-like Mechanism in Arabidopsis. J. Exp. Bot. 2020, 71, 3157–3171. [Google Scholar] [CrossRef]
- Gautier, A.T.; Chambaud, C.; Brocard, L.; Ollat, N.; Gambetta, G.A.; Delrot, S.; Cookson, S.J. Merging Genotypes: Graft Union Formation and Scion-Rootstock Interactions. J. Exp. Bot. 2019, 70, 805–815. [Google Scholar] [CrossRef]
- Tietel, Z.; Srivastava, S.; Fait, A.; Tel-zur, N.; Carmi, N.; Id, E.R. Impact of scion/rootstock reciprocal effects on metabolomics of fruit juice and phloem sap in grafted Citrus reticulata. PLoS ONE 2020, 15, e0227192. [Google Scholar] [CrossRef]
- Macaisne, N.; Novatchkova, M.; Peirera, L.; Vezon, D.; Jolivet, S.; Froger, N.; Chelysheva, L.; Grelon, M.; Mercier, R. SHOC1, an XPF Endonuclease-Related Protein, Is Essential for the Formation of Class I Meiotic Crossovers. Curr. Biol. 2008, 18, 1432–1437. [Google Scholar] [CrossRef]
- An, J.; Guo, Z.; Gou, X.; Li, J. TCP1 positively regulates the expression of DWF4 in arabidopsis thaliana. Plant Signal Behav. 2011, 6, 1117–1118. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, J.; Peng, W.; Peng, D.; Zhuo, Y.; Zhu, D.; Huang, X.; Tang, D.; Guo, M.; He, R.; et al. Dwarfism in Brassica Napus L. Induced by the over-Expression of a Gibberellin 2-Oxidase Gene from Arabidopsis Thaliana. Mol. Breed. 2012, 29, 115–127. [Google Scholar] [CrossRef]
- Yu, L.H.; Miao, Z.Q.; Qi, G.F.; Wu, J.; Cai, X.T.; Mao, J.L.; Xiang, C. Bin MADS-Box Transcription Factor AGL21 Regulates Lateral Root Development and Responds to Multiple External and Physiological Signals. Mol. Plant 2014, 7, 1653–1669. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, S.; Wang, Y.; Zhang, X.; Lv, B.; Luo, L.; Xi, D.; Shen, J.; Ma, H.; Ming, F. OsNAC2 Encoding a NAC Transcription Factor That Affects Plant Height through Mediating the Gibberellic Acid Pathway in Rice. Plant J. 2015, 82, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Panigrahi, K.C. GIGANTEA—An Emerging Story. Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Betsch, L.; Savarin, J.; Bendahmane, M.; Szecsi, J. Roles of the Translationally Controlled Tumor Protein (TCTP) in Plant Development. In Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2017; Volume 64, pp. 149–172. [Google Scholar]
- Liu, Y.; Dong, Q.; Kita, D.; Huang, J.B.; Liu, G.; Wu, X.; Zhu, X.; Cheung, A.Y.; Wu, H.M.; Tao, L.Z. RopGEF1 Plays a Critical Role in Polar Auxin Transport in Early Development. Plant Physiol. 2017, 175, 157–171. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, S.; Li, P.; Yin, Y.; Niu, Q.; Yan, J.; Huang, D. Plant Buffering against the High-Light Stress-Induced Accumulation of CsGA2ox8 Transcripts via Alternative Splicing to Finely Tune Gibberellin Levels and Maintain Hypocotyl Elongation. Hortic. Res. 2021, 8, 2. [Google Scholar] [CrossRef]
- Zhuang, Y.; Wei, M.; Ling, C.; Liu, Y.; Amin, A.K.; Li, P.; Li, P.; Hu, X.; Bao, H.; Huo, H.; et al. EGY3 Mediates Chloroplastic ROS Homeostasis and Promotes Retrograde Signaling in Response to Salt Stress in Arabidopsis. Cell Rep. 2021, 36, 109384. [Google Scholar] [CrossRef]
- Sugimoto, H.; Tanaka, T.; Muramoto, N.; Kitagawa-Yogo, R.; Mitsukawa, N. Transcription Factor NTL9 Negatively Regulates Arabidopsis Vascular Cambium Development during Stem Secondary Growth. Plant Physiol. 2022, 190, 1731–1746. [Google Scholar] [CrossRef]
- Neff, M.M.; Nguyen, S.M.; Malancharuvil, E.J.; Fujioka, S.; Noguchi, T.; Seto, H.; Tsubuki, M.; Honda, T.; Takatsuto, S.; Yoshida, S.; et al. Bas1: A Gene Regulating Brassinosteroid Levels and Light Responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 15316–15323. [Google Scholar] [CrossRef]
- Turk, E.M.; Fujioka, S.; Seto, H.; Shimada, Y.; Takatsuto, S.; Yoshida, S.; Wang, H.; Torres, Q.I.; Ward, J.M.; Murthy, G.; et al. BAS1 and SOB7 Act Redundantly to Modulate Arabidopsis Photomorphogenesis via Unique Brassinosteroid Inactivation Mechanisms. Plant J. 2005, 42, 23–34. [Google Scholar] [CrossRef]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 Transcription Factor Activates the Expression of LSH4 and LSH3, Two Members of the ALOG Gene Family, in Shoot Organ Boundary Cells. Plant J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant Cell Wall Extensibility: Connecting Plant Cell Growth with Cell Wall Structure, Mechanics, and the Action of Wall-Modifying Enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, T.; Xu, B.; Jia, L.; Xiao, B.; Liu, H.; Liu, L. CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase 8 (CCD8) in Tobacco Affects Shoot and Root Architecture. Front. Plant Sci. 2018, 19, 1062. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.; Jiang, H.; Tang, B.; Zhang, M.; Li, Z.; Yin, Y. The AP2/ERF Transcription Factor TINY Modulates Brassinosteroid-Regulated Plant Growth and Drought Responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef] [PubMed]
- Molas, M.L.; Kiss, J.Z. Chapter 1 Phototropism and Gravitropism in Plants. Adv. Bot. Res. 2009, 49, 1–34. [Google Scholar] [CrossRef]
- Yadav, A.; Singh, D.; Lingwan, M.; Yadukrishnan, P.; Masakapalli, S.K.; Datta, S. Light Signaling and UV-B-Mediated Plant Growth Regulation. J. Integr. Plant Biol. 2020, 62, 1270–1292. [Google Scholar] [CrossRef] [PubMed]
- Ledger, S.; Strayer, C.; Ashton, F.; Kay, S.A.; Putterill, J. Analysis of the Function of Two Circadian-Regulated CONSTANS-LIKE Genes. Plant J. 2001, 26, 15–22. [Google Scholar] [CrossRef]
- Ziemienowicz, A.; Haasen, D.; Staiger, D.; Merkle, T. Arabidopsis Transportin1 Is the Nuclear Import Receptor for the Circadian Clock-Regulated RNA-Binding Protein AtGRP7. Plant Mol. Biol. 2003, 53, 201–212. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA Complex Formation Is Required for Day-Length Measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Chia, T.Y.P.; Müller, A.; Jung, C.; Mutasa-Göttgens, E.S. Sugar Beet Contains a Large CONSTANS-LIKE Gene Family Including a CO Homologue That Is Independent of the Early-Bolting (B) Gene Locus. J. Exp. Bot. 2008, 59, 2735–2748. [Google Scholar] [CrossRef]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. The Circadian Clock and Auxin Pathways. For. Genet. 2009, 106, 16883–16888. [Google Scholar]
- Jones, M.A.; Harmer, S.L. JMJD5 Functions in Concert with TOC1 in the Arabidopsis Circadian System. Plant Signal. Behav. 2011, 6, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA Directly Activates Flowering Locus T in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct Roles of FKF1, GIGANTEA, and ZEITLUPE Proteins in the Regulation of Constans Stability in Arabidopsis Photoperiodic Flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef]
- Kiełbowicz-Matuk, A.; Czarnecka, J.; Banachowicz, E.; Rey, P.; Rorat, T. Solanum Tuberosum ZPR1 Encodes a Light-Regulated Nuclear DNA-Binding Protein Adjusting the Circadian Expression of StBBX24 to Light Cycle. Plant Cell Environ. 2017, 40, 424–440. [Google Scholar] [CrossRef] [PubMed]
- Salzman, R.A.; Fujita, T.; Zhu-Salzman, K.; Hasegawa, P.M.; Bressan, R.A. An Improved RNA Isolation Method for Plant Tissues Containing High Levels of Phenolic Compounds or Carbohydrates. Plant Mol. Biol. Report. 1999, 17, 11–17. [Google Scholar] [CrossRef]
- Zeng, Y.; Yang, T. RNA Isolation from Highly Viscous Samples Rich in Polyphenols and Polysaccharides. Plant Mol. Biol. Rep. 2002, 20, 5223210. [Google Scholar] [CrossRef]
- Meisel, L.; Fonseca, B.; González, S.; Baeza-Yates, R.; Cambiazo, V.; Campos, R.; Gonzalez, M.; Orellana, A.; Retamales, J.; Silva, H. A Rapid and Efficient Method for Purifying High Quality Total RNA from Peaches (Prunus Persica) for Functional Genomics Analyses. Biol. Res. 2005, 38, 83–88. [Google Scholar] [CrossRef]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Team, G. Manipulation of FASTQ Data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Fernández i Martí, Á.; Frias, L.; et al. Transposons Played a Major Role in the Diversification between the Closely Related Almond and Peach Genomes: Results from the Almond Genome Sequence. Plant J. 2020, 101, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Marcel, E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-Sequencing Analysis Workflows Using Whole-Transcriptome RT-QPCR Expression Data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef]
- Coenye, T. Do results obtained with RNA-sequencing require independent verification? Biofilm 2021, 3, 100043. [Google Scholar] [CrossRef]


| Cultivar | Rootstock | Length (mm) | d_Scion (mm) | d_Rootstock (mm) |
|---|---|---|---|---|
| ‘Isabelona’ | ‘GN-8’ | 210 a | 2.63 a | 4.25 a |
| Garnem® | 260 b | 3.25 ab | 4.36 a | |
| ‘Lauranne’ | ‘GN-8’ | 310 c | 2.97 ab | 4.50 a |
| Garnem® | 400 d | 3.32 b | 4.56 a |
| logFC | P. dulcis ID | Gene | GO Term | Biological Process |
|---|---|---|---|---|
| 1.392 | Prudul26A014401 | AGL62 | GO:2000012 | regulation of auxin polar transport |
| 1.095 | Prudul26A014156 | ATX1 | GO:0009737 | response to abscisic acid |
| 1.509 | Prudul26A001827 | BPG2 | GO:0009742 | Brassinosteroid-mediated signaling pathway |
| −1.803 | Prudul26A026577 | BRC1 | GO:2000032 | regulation of secondary shoot formation |
| 0.786 | Prudul26A031863 | BZR1 | GO:0009742 | Brassinosteroid-mediated signaling pathway |
| −1.060 | Prudul26A020544 | COL2 | GO:2000028 | regulation of photoperiodism, flowering |
| −1.319 | Prudul26A006124 | DRM1 | GO:0006346 | DNA methylation-dependent heterochromatin formation |
| −0.796 | Prudul26A027812 | DWF4 | GO:0009741 | response to brassinosteroid |
| 3.795 | Prudul26A014067 | ERF106 | GO:0009873 | ethylene-activated signaling pathway |
| −0.833 | Prudul26A003763 | GA3OX1 | GO:0009739 | response to gibberellin |
| 1.069 | Prudul26A008575 | GASA10 | GO:0009740 | gibberellic acid-mediated signaling pathway |
| −1.422 | Prudul26A017626 | GH3.6 | GO:0009733 | response to auxin |
| −0.993 | Prudul26A009430 | HVA22D | GO:0009737 | response to abscisic acid |
| −0.850 | Prudul26A024452 | IAA4 | GO:0009733 | response to auxin |
| −0.793 | Prudul26A000470 | ICK5 | GO:0007049 | cell cycle |
| 0.938 | Prudul26A032509 | KAO2 | GO:0009686 | gibberellin biosynthetic process |
| −1.225 | Prudul26A009950 | KING1 | GO:0042128 | nitrate assimilation |
| −1.390 | Prudul26A008149 | LBD38 | GO:0010468 | regulation of gene expression |
| −1.107 | Prudul26A011380 | LSB1 | GO:0006865 | amino acid transport |
| 0.913 | Prudul26A016769 | MES17 | GO:0048367 | shoot system development |
| −1.064 | Prudul26A032130 | MYB3 | GO:0009800 | cinnamic acid biosynthetic process |
| −0.953 | Prudul26A007232 | NRT1 | GO:0042128 | nitrate assimilation |
| −1.290 | Prudul26A015004 | NRT1 | GO:0042128 | nitrate assimilation |
| 1.409 | Prudul26A017899 | PDX1 | GO:0030154 | cell differentiation |
| −1.687 | Prudul26A015061 | PE11 | GO:0042545 | cell wall modification |
| −1.601 | Prudul26A031322 | PERK4 | GO:0009738 | abscisic acid-activated signaling pathway |
| 0.780 | Prudul26A001835 | PME51 | GO:0042545 | cell wall modification |
| 1.440 | Prudul26A005193 | RALFL34 | GO:0019722 | calcium-mediated signaling |
| 1.179 | Prudul26A030616 | RAP2.3 | GO:0009873 | ethylene-activated signaling pathway |
| −1.154 | Prudul26A000867 | RAP2.7 | GO:0009873 | ethylene-activated signaling pathway |
| −0.967 | Prudul26A017144 | RVE1 | GO:0009734 | auxin-activated signaling pathway |
| −1.305 | Prudul26A019438 | RVE7 | GO:0007623 | circadian rhythm |
| −1.080 | Prudul26A001965 | SGT1 | GO:0009734 | auxin-activated signaling pathway |
| 0.763 | Prudul26A008234 | SKU5 | GO:0009932 | cell tip growth |
| 0.815 | Prudul26A029219 | SRT2 | GO:0009873 | ethylene-activated signaling pathway |
| 0.801 | Prudul26A019903 | SWI3B | GO:0006338 | chromatin remodeling |
| −0.828 | Prudul26A006961 | ZFP7 | GO:0009738 | abscisic acid-activated signaling pathway |
| logFC | P. dulcis ID | Gene | GO Term | Biological Process |
|---|---|---|---|---|
| −1.418 | Prudul26A015064 | CLV1 | GO:0030154 | cell differentiation |
| −1.732 | Prudul26A007496 | DRMH3 | GO:0048364 | root development |
| −0.767 | Prudul26A027600 | EF1A | GO:0030154 | cell differentiation |
| −2.116 | Prudul26A020498 | FIP37 | GO:0010073 | meristem maintenance |
| −1.060 | Prudul26A010669 | GRP7 | GO:0007623 | circadian rhythm |
| −0.909 | Prudul26A029624 | IAMT1 | GO:0009851 | auxin biosynthetic process |
| −1.209 | Prudul26A032200 | JAZ3 | GO:2000022 | regulation of jasmonic acid mediated signaling pathway |
| −0.858 | Prudul26A031524 | LAX3 | GO:0009733 | response to auxin |
| −2.079 | Prudul26A001972 | RD22 | GO:0009651 | response to salt stress |
| −1.578 | Prudul26A025497 | TCTP | GO:0051301 | cell division |
| −1.542 | Prudul26A008660 | VIM1 | GO:0051301 | cell division |
| logFC | P. dulcis ID | Gene | GO Term | Biological Process |
|---|---|---|---|---|
| −1.449 | Prudul26A026190 | LTP3 | GO:0009737 | response to abscisic acid |
| 0.761 | Prudul26A003389 | GLR3.7 | GO:0009737 | response to abscisic acid |
| −1.191 | Prudul26A024452 | IAA4 | GO:0009733 | response to auxin |
| −1.679 | Prudul26A006124 | DRM1 | GO:0006346 | DNA methylation-dependent heterochromatin formation |
| −1.688 | Prudul26A025556 | SAUR50 | GO:0009733 | response to auxin |
| −0.818 | Prudul26A024388 | ROPGEF1 | GO:2000012 | regulation of auxin polar transport |
| −1.135 | Prudul26A031524 | LAX3 | GO:2000012 | regulation of auxin polar transport |
| −0.803 | Prudul26A010146 | TINY2 | GO:0009741 | response to brassinosteroid |
| −1.702 | Prudul26A030744 | BAS1 | GO:0009741 | response to brassinosteroid |
| 1.428 | Prudul26A014814 | SHOC1 | GO:0000712 | resolution of meiotic recombination intermediates |
| 1.217 | Prudul26A029769 | MCM4 | GO:1902969 | mitotic DNA replication |
| 1.192 | Prudul26A003343 | RCC1 | GO:0051301 | cell division |
| 1.121 | Prudul26A018495 | RCC1 | GO:0051301 | cell division |
| 0.932 | Prudul26A013222 | MCM6 | GO:1902969 | mitotic DNA replication |
| 0.779 | Prudul26A022273 | PUR4 | GO:0006541 | glutamine metabolic process |
| −1.120 | Prudul26A008660 | VIM1 | GO:0051301 | cell division |
| 1.657 | Prudul26A020211 | 4CLL6 | GO:0042545 | cell wall modification |
| 0.828 | Prudul26A010546 | GALT1 | GO:0006486 | protein glycosylation |
| −0.964 | Prudul26A003244 | CYT1 | GO:0030244 | cellulose biosynthetic process |
| −1.009 | Prudul26A022981 | NST1 | GO:0009834 | plant-type secondary cell wall biogenesis |
| 1.199 | Prudul26A005611 | HSFB2B | GO:0071456 | cellular response to hypoxia |
| 0.782 | Prudul26A002278 | ZPR1 | GO:0010358 | leaf shaping |
| −0.866 | Prudul26A022967 | CAT2 | GO:0009416 | response to light stimulus |
| −1.000 | Prudul26A017144 | RVE1 | GO:0009734 | auxin-activated signaling pathway |
| −1.102 | Prudul26A014609 | JMJD5 | GO:0007623 | circadian rhythm |
| −1.196 | Prudul26A019438 | RVE7 | GO:0007623 | circadian rhythm |
| −1.227 | Prudul26A010669 | GRP7 | GO:0007623 | circadian rhythm |
| −3.776 | Prudul26A032739 | GRP7 | GO:0007623 | circadian rhythm |
| −1.784 | Prudul26A017801 | CKX5 | GO:0009823 | cytokinin catabolic process |
| 1.943 | Prudul26A021691 | EGY3 | GO:0009651 | response to salt stress |
| 1.551 | Prudul26A019984 | ERF024 | GO:0009873 | ethylene-activated signaling pathway |
| −0.871 | Prudul26A022693 | RCE1 | GO:0009733 | response to auxin |
| 2.056 | Prudul26A000689 | GA2OX8 | GO:0009686 | gibberellin biosynthetic process |
| −1.423 | Prudul26A003763 | GA3OX1 | GO:0009739 | response to gibberellin |
| −1.057 | Prudul26A032482 | NAC2 | GO:0009644 | response to high light intensity |
| −1.141 | Prudul26A032200 | JAZ3 | GO:2000022 | regulation of jasmonic acid mediated signaling pathway |
| −0.982 | Prudul26A012618 | RPT2 | GO:0009638 | phototropism |
| −1.180 | Prudul26A007555 | CDF3 | GO:0009908 | flower development |
| −1.273 | Prudul26A009950 | KING1 | GO:0042128 | nitrate assimilation |
| 1.801 | Prudul26A016707 | GI | GO:0030154 | cell differentiation |
| 2.142 | Prudul26A024652 | PAT1 | GO:0009640 | photomorphogenesis |
| 1.422 | Prudul26A014018 | NTL9 | GO:0071470 | cellular response to osmotic stress |
| 0.782 | Prudul26A016310 | TOE3 | GO:0009873 | ethylene-activated signaling pathway |
| −0.763 | Prudul26A027253 | GCR2 | GO:0005975 | carbohydrate metabolic process |
| −0.769 | Prudul26A006961 | ZFP7 | GO:0009738 | abscisic acid-activated signaling pathway |
| −0.820 | Prudul26A024443 | COL4 | GO:0009909 | regulation of flower development |
| −0.919 | Prudul26A002088 | SUI1 | GO:0030154 | cell differentiation |
| −0.994 | Prudul26A002057 | LSH3 | GO:0010492 | maintenance of shoot apical meristem identity |
| −1.234 | Prudul26A016351 | CDF2 | GO:0009908 | flower development |
| −1.264 | Prudul26A025497 | TCTP | GO:0051301 | cell division |
| −1.291 | Prudul26A015967 | SPL9 | GO:0048366 | leaf development |
| −1.586 | Prudul26A020498 | FIP37 | GO:0010073 | meristem maintenance |
| −1.971 | Prudul26A022418 | MAX1 | GO:0009926 | auxin polar transport |
| 0.864 | Prudul26A016413 | AGL21 | GO:0048364 | root development |
| −1.010 | Prudul26A029350 | JKD | GO:0048364 | root development |
| −1.082 | Prudul26A020939 | AGL79 | GO:0010311 | lateral root formation |
| −1.674 | Prudul26A007496 | DRMH3 | GO:0048364 | root development |
| 1.705 | Prudul26A006492 | SWEET2 | GO:0008643 | carbohydrate transport |
| −0.983 | Prudul26A013154 | SWEET2 | GO:0008643 | carbohydrate transport |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montesinos, Á.; Rubio-Cabetas, M.J.; Grimplet, J. Characterization of Almond Scion/Rootstock Communication in Cultivar and Rootstock Tissues through an RNA-Seq Approach. Plants 2023, 12, 4166. https://doi.org/10.3390/plants12244166
Montesinos Á, Rubio-Cabetas MJ, Grimplet J. Characterization of Almond Scion/Rootstock Communication in Cultivar and Rootstock Tissues through an RNA-Seq Approach. Plants. 2023; 12(24):4166. https://doi.org/10.3390/plants12244166
Chicago/Turabian StyleMontesinos, Álvaro, María José Rubio-Cabetas, and Jérôme Grimplet. 2023. "Characterization of Almond Scion/Rootstock Communication in Cultivar and Rootstock Tissues through an RNA-Seq Approach" Plants 12, no. 24: 4166. https://doi.org/10.3390/plants12244166
APA StyleMontesinos, Á., Rubio-Cabetas, M. J., & Grimplet, J. (2023). Characterization of Almond Scion/Rootstock Communication in Cultivar and Rootstock Tissues through an RNA-Seq Approach. Plants, 12(24), 4166. https://doi.org/10.3390/plants12244166








