Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses
Abstract
1. Introduction
2. Classification of RLKs
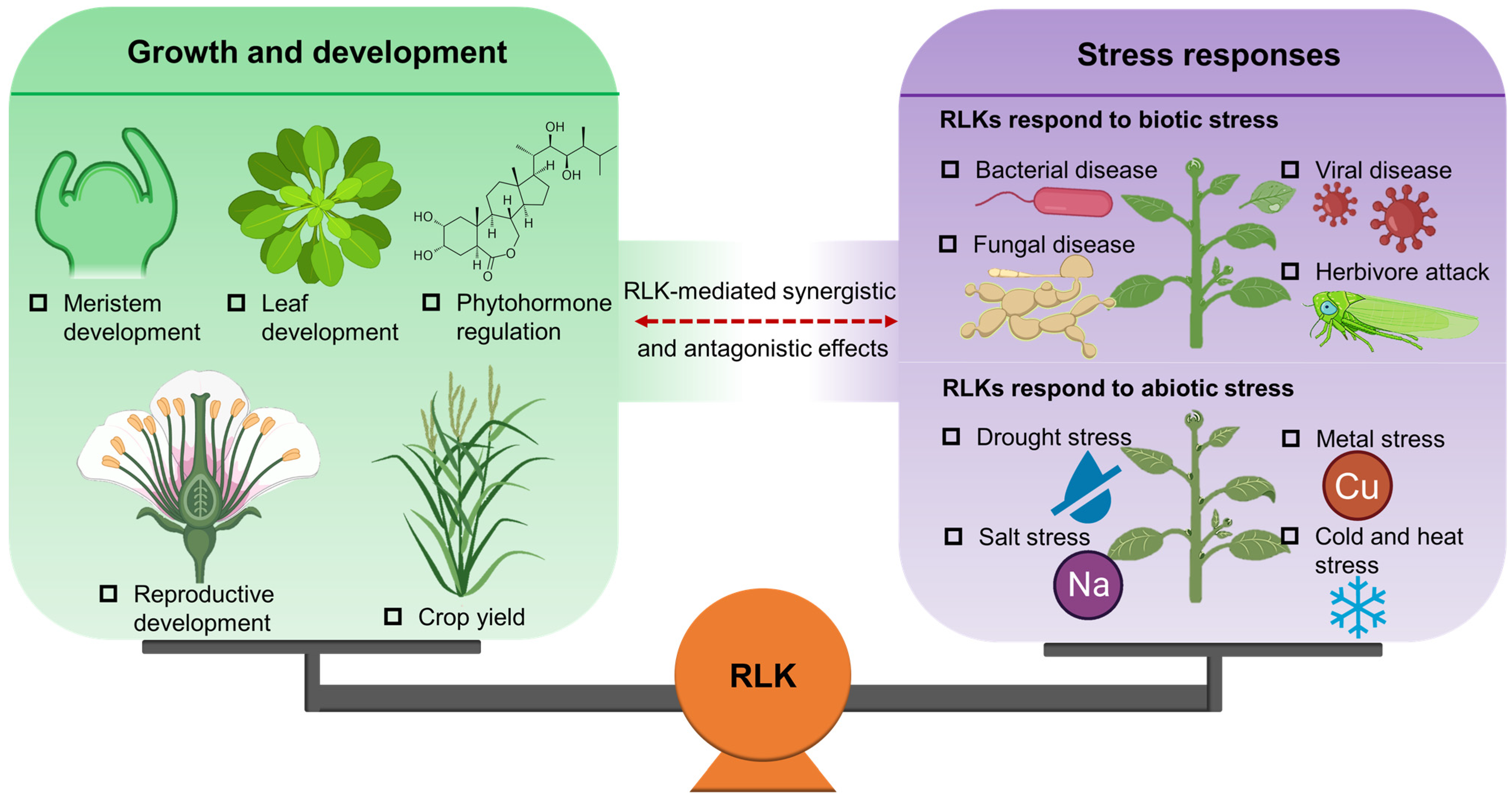
3. Regulation of RLKs on Plant Growth and Development
3.1. Meristem Development
3.2. Leaf Development
3.3. Reproductive Development
3.4. Crop Yield
3.5. Phytohormone Regulation
4. Biological Functions of RLKs in Plant Stress Response
4.1. RLKs Respond to Biotic Stress
4.1.1. Bacterial Disease
4.1.2. Fungal Diseases
4.1.3. Viral Disease
4.1.4. Herbivore Attack
4.2. RLKs Respond to Abiotic Stress
4.2.1. Drought Stress
4.2.2. Salt Stress
4.2.3. Metal Stress
4.2.4. Cold and Heat Stress
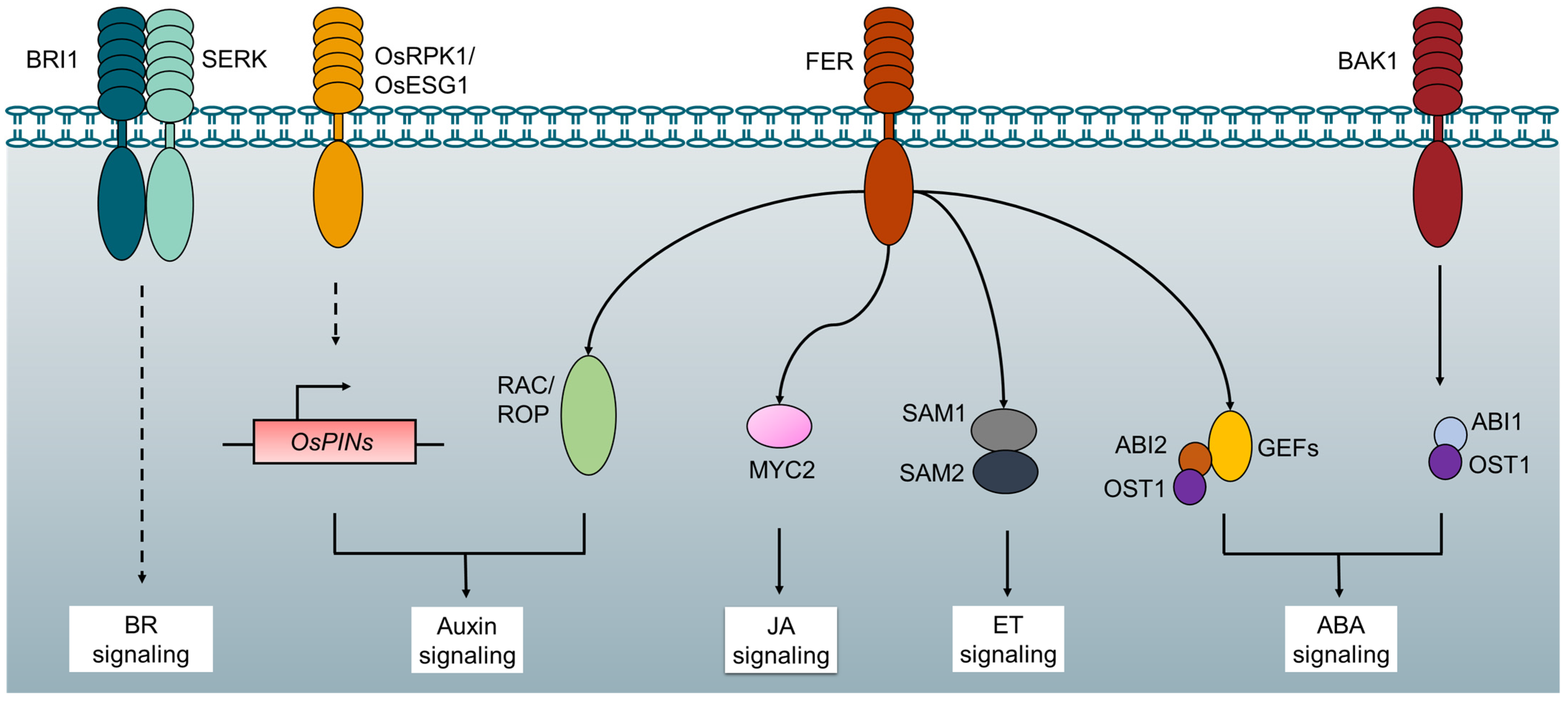
5. RLK-Mediated Molecular Crosstalk between Plant Growth and Stress Response
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, D.H.; Zaman, W.; Lu, J.; Niu, Q.; Zhang, X.; Ayaz, A.; Saqib, S.; Yang, B.; Zhang, J.; Zhao, H.; et al. Natural lupeol level variation among castor accessions and the upregulation of lupeol synthesis in response to light. Ind. Crops Prod. 2023, 192, 116090. [Google Scholar] [CrossRef]
- Walker, J.C.; Zhang, R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of brassica. Nature 1990, 345, 743–746. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Escocard de Azevedo Manhães, A.M.; Ortiz-Morea, F.A.; He, P.; Shan, L. Plant plasma membrane-resident receptors: Surveillance for infections and coordination for growth and development. J. Integr. Plant Biol. 2021, 63, 79–101. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.S.; De Britto, S.; Nagaraj, G.; Gurulingaiah, B.; Shekhar, R.; Ito, S.-i.; Jogaiah, S. Genome-Wide Identification, Diversification, and Expression Analysis of Lectin Receptor-Like Kinase (LecRLK) Gene Family in Cucumber under Biotic Stress. Int. J. Mol. Sci. 2021, 22, 6585. [Google Scholar] [CrossRef] [PubMed]
- Soltabayeva, A.; Dauletova, N.; Serik, S.; Sandybek, M.; Omondi, J.O.; Kurmanbayeva, A.; Srivastava, S. Receptor-like Kinases (LRR-RLKs) in Response of Plants to Biotic and Abiotic Stresses. Plants 2022, 11, 2660. [Google Scholar] [CrossRef]
- Abedi, A.; Hajiahmadi, Z.; Kordrostami, M.; Esmaeel, Q.; Jacquard, C. Analyses of Lysin-motif Receptor-like Kinase (LysM-RLK) Gene Family in Allotetraploid Brassica napus L. and Its Progenitor Species: An In Silico Study. Cells 2022, 11, 37. [Google Scholar] [CrossRef]
- Jose, J.; Ghantasala, S.; Choudhury, S.R. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef]
- Mishra, D.; Suri, G.S.; Kaur, G.; Tiwari, M. Comprehensive analysis of structural, functional, and evolutionary dynamics of Leucine Rich Repeats-RLKs in Thinopyrum elongatum. Int. J. Biol. Macromol. 2021, 183, 513–527. [Google Scholar] [CrossRef]
- Dievart, A.; Gottin, C.; Perin, C.; Ranwez, V.; Chantret, N. Origin and Diversity of Plant Receptor-Like Kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef]
- Monaghan, J. Conserved Degradation of Orthologous RLCKs Regulates Immune Homeostasis. Trends Plant Sci. 2018, 23, 554–557. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Zipfel, C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef]
- Takahashi, F.; Shinozaki, K. Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 2019, 47, 106–111. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Balcerowicz, M.; Di Antonio, M.; Jaeger, K.E.; Geng, F.; Franaszek, K.; Marriott, P.; Brierley, I.; Firth, A.E.; Wigge, P.A. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 2020, 6, 522–532. [Google Scholar] [CrossRef]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar] [CrossRef]
- Clark, S.E.; Williams, R.W.; Meyerowitz, E.M. The CLAVATA1Gene Encodes a Putative Receptor Kinase That Controls Shoot and Floral Meristem Size in Arabidopsis. Cell 1997, 89, 575–585. [Google Scholar] [CrossRef]
- Cui, Y.W.; Hu, C.; Zhu, Y.F.; Cheng, K.L.; Li, X.N.; Wei, Z.Y.; Xue, L.; Lin, F.; Shi, H.Y.; Yi, J.; et al. CIK Receptor Kinases Determine Cell Fate Specification during Early Anther Development in Arabidopsis. Plant Cell 2018, 30, 2383–2401. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef] [PubMed]
- Gujas, B.; Kastanaki, E.; Sturchler, A.; Cruz, T.M.D.; Ruiz-Sola, M.A.; Dreos, R.; Eicke, S.; Truernit, E.; Rodriguez-Villalon, A. A Reservoir of Pluripotent Phloem Cells Safeguards the Linear Developmental Trajectory of Protophloem Sieve Elements. Curr. Biol. 2020, 30, 755–766.e4. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Fujihara, R.; Uchida, N.; Tameshige, T.; Kawamoto, N.; Hotokezaka, Y.; Higaki, T.; Simon, R.; Torii, K.U.; Tasaka, M.; Aida, M. The boundary-expressed EPIDERMAL PATTERNING FACTOR-LIKE2 gene encoding a signaling peptide promotes cotyledon growth during Arabidopsis thaliana embryogenesis. Plant Biotechnol. 2021, 38, 317–322. [Google Scholar] [CrossRef]
- Kawamoto, N.; Del Carpio, D.P.; Hofmann, A.; Mizuta, Y.; Kurihara, D.; Higashiyama, T.; Uchida, N.; Torii, K.U.; Colombo, L.; Groth, G.; et al. A Peptide Pair Coordinates Regular Ovule Initiation Patterns with Seed Number and Fruit Size. Curr. Biol. 2020, 30, 4352–4361.e4. [Google Scholar] [CrossRef]
- Liu, T.; Jiang, G.Q.; Yao, X.F.; Liu, C.M. The leucine-rich repeat receptor-like kinase OsERL plays a critical role in anther lobe formation in rice. Biochem. Biophys. Res. Commun. 2021, 563, 85–91. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, R.; Gui, J.S.; Zhong, Y.; Li, L.G. The Receptor-Like Kinase AtVRLK1 Regulates Secondary Cell Wall Thickening. Plant Physiol. 2018, 177, 671–683. [Google Scholar] [CrossRef]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Mangano, S.; Marzol, E.; Denita-Juarez, S.P.; Salgado Salter, J.D.; Velasquez, S.M.; Muschietti, J.P.; Estevez, J.M. Proline-rich extensin-like receptor kinases PERK5 and PERK12 are involved in pollen tube growth. FEBS Lett. 2021, 595, 2593–2607. [Google Scholar] [CrossRef]
- Yu, J.P.; Han, J.J.; Kim, Y.J.; Song, M.; Yang, Z.; He, Y.F.; Fu, R.F.; Luo, Z.J.; Hu, J.P.; Liang, W.Q.; et al. Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc. Natl. Acad. Sci. USA 2017, 114, 12327–12332. [Google Scholar] [CrossRef]
- Wang, B.; Fang, R.Q.; Zhang, J.; Han, J.L.; Chen, F.M.; He, F.R.; Liu, Y.G.; Chen, L.T. Rice LecRK5 phosphorylates a UGPase to regulate callose biosynthesis during pollen development. J. Exp. Bot. 2020, 71, 4033–4041. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.H.; Qin, Z.R.; Zhang, C.Y.; Liu, B.; Liu, J.; Zhang, C.S.; Lin, C.T.; Li, H.Y.; Zhao, T. Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J. Exp. Bot. 2015, 66, 7197–7209. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Lu, Z.Q.; Shan, J.X.; Ye, W.W.; Dong, N.Q.; Lin, H.X. ERECTA1 Acts Upstream of the OsMKKK10-OsMKK4-OsMPK6 Cascade to Control Spikelet Number by Regulating Cytokinin Metabolism in Rice. Plant Cell 2020, 32, 2763–2779. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Breja, P.; Khurana, J.P.; Khurana, P. Wheat Brassinosteroid-Insensitive1 (TaBRI1) Interacts with Members of TaSERK Gene Family and Cause Early Flowering and Seed Yield Enhancement in Arabidopsis. PLoS ONE 2016, 11, e0153273. [Google Scholar] [CrossRef]
- Sharma, N.; Khurana, P. Genome-wide identification, characterization and expression analysis of the BRI1 gene family in Triticum aestivum L. Plant Biotechnol. Rep. 2022, 16, 777–791. [Google Scholar] [CrossRef]
- Rafeie, M.; Amerian, M.R.; Sorkhi, B.; Heidari, P.; Asghari, H.R. Effect of Exogenous Brassinosteroid Application on Grain Yield, some Physiological Traits and Expression of Genes Related to This Hormone Signaling Pathway in Wheat under Drought Stress. Plant Genet. Res. 2020, 6, 157–172. [Google Scholar] [CrossRef]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A Brassinosteroid-lnsensitive Mutant in Arabidopsis thaliana Exhibits Multiple Defects in Growth and Development. Plant Physiol. 1996, 3111, 671–678. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef]
- Lozano-Elena, F.; Caño-Delgado, A.I. Emerging roles of vascular brassinosteroid receptors of the BRI1-like family. Curr. Opin. Plant Biol. 2019, 51, 105–113. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V. Linking brassinosteroid and ABA signaling in the context of stress acclimation. Int. J. Mol. Sci. 2020, 21, 5108. [Google Scholar] [CrossRef]
- Kim, S.Y.; Warpeha, K.M.; Huber, S.C. The brassinosteroid receptor kinase, BRI1, plays a role in seed germination and the release of dormancy by cold stratification. J. Plant Physiol. 2019, 241, 153031. [Google Scholar] [CrossRef]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis Somatic Embryogenesis Receptor Kinase 1 Gene Is Expressed in Developing Ovules and Embryos and Enhances Embryogenic Competence in Culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef]
- Hohmann, U.; Ramakrishna, P.; Wang, K.; Lorenzo-Orts, L.; Nicolet, J.; Henschen, A.; Barberon, M.; Bayer, M.; Hothorn, M. Constitutive Activation of Leucine-Rich Repeat Receptor Kinase Signaling Pathways by BAK1-INTERACTING RECEPTOR-LIKE KINASE3 Chimera. Plant Cell 2020, 32, 3311–3323. [Google Scholar] [CrossRef]
- Ackerman-Lavert, M.; Savaldi-Goldstein, S. Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 2020, 53, 90–97. [Google Scholar] [CrossRef]
- Chen, W.Y.; Lv, M.H.; Wang, Y.Z.; Wang, P.A.; Cui, Y.W.; Li, M.Z.; Wang, R.S.; Gou, X.P.; Li, J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 2019, 10, 4164. [Google Scholar] [CrossRef]
- Zheng, B.W.; Bai, Q.W.; Wu, L.; Liu, H.; Liu, Y.P.; Xu, W.J.; Li, G.S.; Ren, H.Y.; She, X.P.; Wu, G. EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 2019, 10, 4165. [Google Scholar] [CrossRef]
- Dong, N.N.; Yin, W.C.; Liu, D.P.; Zhang, X.X.; Yu, Z.K.; Huang, W.; Liu, J.H.; Yang, Y.Z.; Meng, W.J.; Niu, M.; et al. Regulation of Brassinosteroid Signaling and Salt Resistance by SERK2 and Potential Utilization for Crop Improvement in Rice. Front. Plant Sci. 2020, 11, 621859. [Google Scholar] [CrossRef]
- Hajný, J.; Prát, T.; Rydza, N.; Rodriguez, L.; Tan, S.; Verstraeten, I.; Domjan, D.; Mazur, E.; Smakowska-Luzan, E.; Smet, W. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 2020, 370, 550–557. [Google Scholar] [CrossRef]
- Marques-Bueno, M.M.; Armengot, L.; Noack, L.C.; Bareille, J.; Rodriguez, L.; Platre, M.P.; Bayle, V.; Liu, M.; Opdenacker, D.; Vanneste, S. Auxin-regulated reversible inhibition of TMK1 signaling by MAKR2 modulates the dynamics of root gravitropism. Curr. Biol. 2021, 31, 228–237.e210. [Google Scholar] [CrossRef]
- Cammarata, J.; Morales Farfan, C.; Scanlon, M.J.; Roeder, A.H.K. Cytokinin–CLAVATA cross-talk is an ancient mechanism regulating shoot meristem homeostasis in land plants. Proc. Natl. Acad. Sci. USA 2022, 119, e2116860119. [Google Scholar] [CrossRef]
- Stahl, E.; Martin, A.F.; Glauser, G.; Guillou, M.-C.; Aubourg, S.; Renou, J.-P.; Reymond, P. The MIK2/SCOOP signaling system contributes to Arabidopsis resistance against herbivory by modulating jasmonate and indole glucosinolate biosynthesis. Front. Plant Sci. 2022, 13, 852808. [Google Scholar] [CrossRef] [PubMed]
- Konopka-Postupolska, D.; Dobrowolska, G. ABA perception is modulated by membrane receptor-like kinases. J. Exp. Bot. 2020, 71, 1210–1214. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fernie, A.R. Remote Control of Transpiration via ABA. Trends Plant Sci. 2018, 23, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.-K.; Takahashi, Y.; Merilo, E.; Costa, A.; Zhang, L.; Kernig, K.; Lee, K.H.; Schroeder, J.I. Raf-like kinases and receptor-like (pseudo)kinase GHR1 are required for stomatal vapor pressure difference response. Proc. Natl. Acad. Sci. USA 2021, 118, e2107280118. [Google Scholar] [CrossRef] [PubMed]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef]
- Thapa, G.; Gunupuru, L.R.; Hehir, J.G.; Kahla, A.; Mullins, E.; Doohan, F.M. A Pathogen-Responsive Leucine Rich Receptor Like Kinase Contributes to Fusarium Resistance in Cereals. Front. Plant Sci. 2018, 9, 867. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef]
- Zou, Y.; Liu, X.Y.; Wang, Q.; Chen, Y.; Liu, C.; Qiu, Y.; Zhang, W. OsRPK1, a novel leucine-rich repeat receptor-like kinase, negatively regulates polar auxin transport and root development in rice. Biochim. Biophys. Acta 2014, 1840, 1676–1685. [Google Scholar] [CrossRef]
- Pan, J.W.; Li, Z.; Wang, Q.G.; Yang, L.Q.; Yao, F.Y.; Liu, W. An S-domain receptor-like kinase, OsESG1, regulates early crown root development and drought resistance in rice. Plant Sci. 2020, 290, 110318. [Google Scholar] [CrossRef]
- Shang, Y.; Dai, C.; Lee, M.M.; Kwak, J.M.; Nam, K.H. BRI1-Associated Receptor Kinase 1 Regulates Guard Cell ABA Signaling Mediated by Open Stomata 1 in Arabidopsis. Mol. Plant 2016, 9, 447–460. [Google Scholar] [CrossRef]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef]
- Mao, D.; Yu, F.; Li, J.; Van de Poel, B.; Tan, D.; Li, J.; Liu, Y.; Li, X.; Dong, M.; Chen, L. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in A rabidopsis. Plant Cell Environ. 2015, 38, 2566–2574. [Google Scholar] [CrossRef]
- Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; Xie, Z.; Chen, J.; Schnable, P.S.; Walley, J.W.; Yin, Y. FERONIA Receptor Kinase Contributes to Plant Immunity by Suppressing Jasmonic Acid Signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324.e6. [Google Scholar] [CrossRef]
- Teixeira, R.M.; Ferreira, M.A.; Raimundo, G.A.S.; Loriato, V.A.P.; Reis, P.A.B.; Fontes, E.P.B. Virus perception at the cell surface: Revisiting the roles of receptor-like kinases as viral pattern recognition receptors. Mol. Plant Pathol. 2019, 20, 1196–1202. [Google Scholar] [CrossRef]
- Coleman, A.D.; Maroschek, J.; Raasch, L.; Takken, F.L.W.; Ranf, S.; Hückelhoven, R. The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytol. 2021, 229, 3453–3466. [Google Scholar] [CrossRef]
- Laohavisit, A.; Wakatake, T.; Ishihama, N.; Mulvey, H.; Takizawa, K.; Suzuki, T.; Shirasu, K. Quinone perception in plants via leucine-rich-repeat receptor-like kinases. Nature 2020, 587, 92–97. [Google Scholar] [CrossRef]
- Malukani, K.K.; Ranjan, A.; Hota, S.J.; Patel, H.K.; Sonti, R.V. Dual Activities of Receptor-Like Kinase OsWAKL21.2 Induce Immune Responses1. Plant Physiol. 2020, 183, 1345–1363. [Google Scholar] [CrossRef]
- Yang, Q.J.; Guo, J.H.; Zeng, H.R.; Xu, L.H.; Xue, J.; Xiao, S.; Li, J.F. The receptor-like cytoplasmic kinase CDG1 negatively regulates Arabidopsis pattern-triggered immunity and is involved in AvrRpm1-induced RIN4 phosphorylation. Plant Cell 2021, 33, 1341–1360. [Google Scholar] [CrossRef]
- Ma, X.Y.; Xu, G.Y.; He, P.; Shan, L.B. SERKing Coreceptors for Receptors. Trends Plant Sci. 2016, 21, 1017–1033. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Elmore, J.M.; Creer, A.Y.; Feng, B.M.; Franco, J.Y.; Rufian, J.S. A Cysteine-Rich Protein Kinase Associates with a Membrane Immune Complex and the Cysteine Residues Are Required for Cell Death. Plant Physiol. 2017, 173, 771–787. [Google Scholar] [CrossRef]
- Yoo, Y.; Park, J.C.; Cho, M.H.; Yang, J.; Kim, C.Y.; Jung, K.H.; Jeon, J.S.; An, G.; Lee, S.W. Lack of a Cytoplasmic RLK, Required for ROS Homeostasis, Induces Strong Resistance to Bacterial Leaf Blight in Rice. Front. Plant Sci. 2018, 9, 577. [Google Scholar] [CrossRef]
- Wang, L.L.; Ma, Z.B.; Kang, H.X.; Gu, S.; Mukhina, Z.; Wang, C.H.; Wang, H.; Bai, Y.J.; Sui, G.M.; Zheng, W.J.; et al. Cloning and functional analysis of the novel rice blast Resistance gene Pi65 in japonica rice. Theor. Appl. Genet. 2022, 135, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.B.; Bai, P.F.; Ning, Y.S.; Wang, J.Y.; Shi, X.T.; Xiong, Y.H.; Zhang, K.; He, F.; Zhang, C.Y.; Wang, R.Y.; et al. The Monocot-Specific Receptor-like Kinase SDS2 Controls Cell Death and Immunity in Rice. Cell Host Microbe 2018, 23, 498–510.e5. [Google Scholar] [CrossRef] [PubMed]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, L.; Wang, P.; Zhang, S.; Wu, J. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC Genom. 2017, 18, 763. [Google Scholar] [CrossRef] [PubMed]
- van der Burgh, A.M.; Postma, J.; Robatzek, S.; Joosten, M. Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 2019, 20, 410–422. [Google Scholar] [CrossRef]
- Wang, J.H.; Wang, J.J.; Shang, H.S.; Chen, X.M.; Xu, X.M.; Hu, X.P. TaXa21, a Leucine-Rich Repeat Receptor-Like Kinase Gene Associated with TaWRKY76 and TaWRKY62, Plays Positive Roles in Wheat High-Temperature Seedling Plant Resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2019, 32, 1526–1535. [Google Scholar] [CrossRef]
- Wang, J.H.; Wang, J.J.; Li, J.; Shang, H.S.; Chen, X.M.; Hu, X.P. The RLK protein TaCRK10 activates wheat high-temperature seedling-plant resistance to stripe rust through interacting with TaH2A.1. Plant J. 2021, 108, 1241–1255. [Google Scholar] [CrossRef]
- Zhang, H.H.; Chen, C.H.; Li, L.L.; Tan, X.X.; Wei, Z.Y.; Li, Y.J.; Li, J.M.; Yan, F. A rice LRR receptor-like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol. J. 2021, 19, 2319–2332. [Google Scholar] [CrossRef]
- Costa, A.T.; Bravo, J.P.; Krause-Sakate, R.; Maia, I.G. The receptor-like kinase SlSOBIR1 is differentially modulated by virus infection but its overexpression in tobacco has no significant impact on virus accumulation. Plant Cell Rep. 2016, 35, 65–75. [Google Scholar] [CrossRef]
- Tran, P.T.; Citovsky, V. Receptor-like kinase BAM1 facilitates early movement of the Tobacco mosaic virus. Commun. Biol. 2021, 4, 511. [Google Scholar] [CrossRef]
- Li, B.; Ferreira, M.A.; Huang, M.L.; Camargos, L.F.; Yu, X.; Teixeira, R.M.; Carpinetti, P.A.; Mendes, G.C.; Gouveia-Mageste, B.C.; Liu, C.L.; et al. The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 2019, 10, 4996. [Google Scholar] [CrossRef]
- Hamann, E.; Blevins, C.; Franks, S.J.; Jameel, M.I.; Anderson, J.T. Climate change alters plant–herbivore interactions. New Phytol. 2021, 229, 1894–1910. [Google Scholar] [CrossRef]
- Maron, J.L.; Agrawal, A.A.; Schemske, D.W. Plant–herbivore coevolution and plant speciation. Ecology 2019, 100, e02704. [Google Scholar] [CrossRef]
- Zu, P.; Boege, K.; del-Val, E.; Schuman, M.C.; Stevenson, P.C.; Zaldivar-Riverón, A.; Saavedra, S. Information arms race explains plant-herbivore chemical communication in ecological communities. Science 2020, 368, 1377–1381. [Google Scholar] [CrossRef]
- Meisrimler, C.-N.; Allan, C.; Eccersall, S.; Morris, R.J. Interior design: How plant pathogens optimize their living conditions. New Phytol. 2021, 229, 2514–2524. [Google Scholar] [CrossRef]
- Klymiuk, V.; Coaker, G.; Fahima, T.; Pozniak, C.J. Tandem Protein Kinases Emerge as New Regulators of Plant Immunity. Mol. Plant Microbe Interact. 2021, 34, 1094–1102. [Google Scholar] [CrossRef]
- Hu, L.F.; Ye, M.; Kuai, P.; Ye, M.F.; Erb, M.; Lou, Y.G. OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 2018, 219, 1097–1111. [Google Scholar] [CrossRef]
- Ye, M.; Kuai, P.; Hu, L.F.; Ye, M.F.; Sun, H.; Erb, M.; Lou, Y.G. Suppression of a leucine-rich repeat receptor-like kinase enhances host plant resistance to a specialist herbivore. Plant Cell Environ. 2020, 43, 2571–2585. [Google Scholar] [CrossRef]
- Kim, H.; Seomun, S.; Yoon, Y.; Jang, G. Jasmonic Acid in Plant Abiotic Stress Tolerance and Interaction with Abscisic Acid. Agronomy 2021, 11, 1886. [Google Scholar] [CrossRef]
- Kumar, M.; Kesawat, M.S.; Ali, A.; Lee, S.-C.; Gill, S.S.; Kim, H.U. Integration of Abscisic Acid Signaling with Other Signaling Pathways in Plant Stress Responses and Development. Plants 2019, 8, 592. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants. Cells 2021, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Q.; Sheng, P.K.; Tan, J.J.; Chen, X.L.; Lu, G.W.; Ma, W.W.; Heng, Y.Q.; Lin, Q.B.; Zhu, S.S.; Wang, J.L.; et al. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 2015, 66, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Liang, C.C.; Hou, S.G.; Wang, X.; Chen, D.H.; Shen, J.L.; Zhang, W.; Wang, M. The LRR-RLK Protein HSL3 Regulates Stomatal Closure and the Drought Stress Response by Modulating Hydrogen Peroxide Homeostasis. Front. Plant Sci. 2020, 11, 548034. [Google Scholar] [CrossRef]
- Shen, J.L.; Diao, W.Z.; Zhang, L.F.; Acharya, B.R.; Wang, M.; Zhao, X.Y.; Chen, D.H.; Zhang, W. Secreted Peptide PIP1 Induces Stomatal Closure by Activation of Guard Cell Anion Channels in Arabidopsis. Front. Plant Sci. 2020, 11, 1029. [Google Scholar] [CrossRef]
- Chowdhury, R.; Mubassir, M.H.M. How Arabidopsis Receptor-Like Kinase 7 (RLK7) Manifests: Delineating Its Structure and Function. Adv. Agric. 2022, 2022, 4715110. [Google Scholar] [CrossRef]
- Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Merilo, E.; Laanemets, K.; Waadt, R.; Pater, D.; Kollist, H.; Schroeder, J.I. Abscisic acid-independent stomatal CO2 signal transduction pathway and convergence of CO2 and ABA signaling downstream of OST1 kinase. Proc. Natl. Acad. Sci. USA 2018, 115, E9971–E9980. [Google Scholar] [CrossRef]
- Ali, A.; Kim, J.K.; Jan, M.; Khan, H.A.; Khan, I.U.; Shen, M.; Park, J.; Lim, C.J.; Hussain, S.; Baek, D.; et al. Rheostatic Control of ABA Signaling through HOS15-Mediated OST1 Degradation. Mol. Plant 2019, 12, 1447–1462. [Google Scholar] [CrossRef]
- Shang, Y.; Yang, D.M.; Ha, Y.M.; Shin, H.Y.; Nam, K.H. Receptor-like protein kinases RPK1 and BAK1 sequentially form complexes with the cytoplasmic kinase OST1 to regulate ABA-induced stomatal closure. J. Exp. Bot. 2020, 71, 1491–1502. [Google Scholar] [CrossRef]
- Chong, L.; Xu, R.; Ku, L.; Zhu, Y. Beyond stress response: OST1 opening doors for plants to grow. Stress Biol. 2022, 2, 44. [Google Scholar] [CrossRef]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.M.; Li, S.; Wang, K.; Tian, H.R.; Gao, J.F.; Zhao, Q.Z.; Du, C.Q. A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 2020, 296, 110465. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.B.; Liu, C.; Tang, D.Y.; Yan, L.; Wang, D.; Yang, Y.Z.; Gui, J.H.; Zhao, X.Y.; Li, L.G.; Tang, X.D.; et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Lv, Y.Y.; Lei, W.R.; Li, X.; Chen, Y.H.; Zheng, L.Q.; Xia, Y.; Shen, Z.G. Cloning and characterization of the Oryza sativa wall-associated kinase gene OsWAK11 and its transcriptional response to abiotic stresses. Plant Soil 2014, 384, 335–346. [Google Scholar] [CrossRef]
- Trinh, N.N.; Huang, T.L.; Chi, W.C.; Fu, S.F.; Chen, C.C.; Huang, H.J. Chromium stress response effect on signal transduction and expression of signaling genes in rice. Physiol. Plant 2014, 150, 205–224. [Google Scholar] [CrossRef]
- Janská, A.; Marsík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Yang, L.; Wu, K.C.; Gao, P.; Liu, X.J.; Li, G.P.; Wu, Z.J. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 2014, 215–216, 19–28. [Google Scholar] [CrossRef]
- Geng, B.H.; Wang, Q.; Huang, R.S.; Liu, Y.J.; Guo, Z.F.; Lu, S.Y. A novel LRR-RLK (CTLK) confers cold tolerance through regulation on the C-repeat-binding factor pathway, antioxidants, and proline accumulation. Plant J. 2021, 108, 1679–1689. [Google Scholar] [CrossRef]
- Wang, H.; Niu, H.; Liang, M.; Zhai, Y.; Huang, W.; Ding, Q.; Du, Y.; Lu, M. A Wall-Associated Kinase Gene CaWAKL20 From Pepper Negatively Modulates Plant Thermotolerance by Reducing the Expression of ABA-Responsive Genes. Front. Plant Sci. 2019, 10, 591. [Google Scholar] [CrossRef]
- Pelagio-Flores, R.; Muñoz-Parra, E.; Barrera-Ortiz, S.; Ortiz-Castro, R.; Saenz-Mata, J.; Ortega-Amaro, M.A.; Jiménez-Bremont, J.F.; López-Bucio, J. The cysteine-rich receptor-like protein kinase CRK28 modulates Arabidopsis growth and development and influences abscisic acid responses. Planta 2019, 251, 2. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, B.H.; Lim, C.J.; Lim, C.O.; Nam, K.H. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol. Plant. 2010, 138, 191–204. [Google Scholar] [CrossRef]
- Chen, X.W.; Zuo, S.M.; Schwessinger, B.; Chern, M.; Canlas, P.E.; Ruan, D.L.; Zhou, X.G.; Wang, J.; Daudi, A.; Petzold, C.J.; et al. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 2014, 7, 874–892. [Google Scholar] [CrossRef]
- Zuo, S.; Zhou, X.; Chen, M.; Zhang, S.; Schwessinger, B.; Ruan, D.; Yuan, C.; Wang, J.; Chen, X.; Ronald, P.C. OsSERK1 regulates rice development but not immunity to Xanthomonas oryzae pv. oryzae or Magnaporthe oryzae. J. Integr. Plant Biol. 2014, 56, 1179–1192. [Google Scholar] [CrossRef]
- Franck, C.M.; Westermann, J.; Boisson-Dernier, A. Plant Malectin-Like Receptor Kinases: From Cell Wall Integrity to Immunity and Beyond. Annu. Rev. Plant Biol. 2018, 69, 301–328. [Google Scholar] [CrossRef]
- Ge, Z.X.; Bergonci, T.; Zhao, Y.L.; Zou, Y.J.; Du, S.; Liu, M.C.; Luo, X.J.; Ruan, H.; García-Valencia, L.E.; Zhong, S.; et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017, 358, 1596–1600. [Google Scholar] [CrossRef]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P.; et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Chakravorty, D.; Yu, Y.Q.; Assmann, S.M. A kinase-dead version of FERONIA receptor-like kinase has dose-dependent impacts on rosette morphology and RALF1-mediated stomatal movements. FEBS Lett. 2018, 592, 3429–3437. [Google Scholar] [CrossRef]
- Wang, L.; Yang, T.; Lin, Q.L.; Wang, B.Q.; Li, X.; Luan, S.; Yu, F. Receptor kinase FERONIA regulates flowering time in Arabidopsis. BMC Plant Biol. 2020, 20, 26. [Google Scholar] [CrossRef]
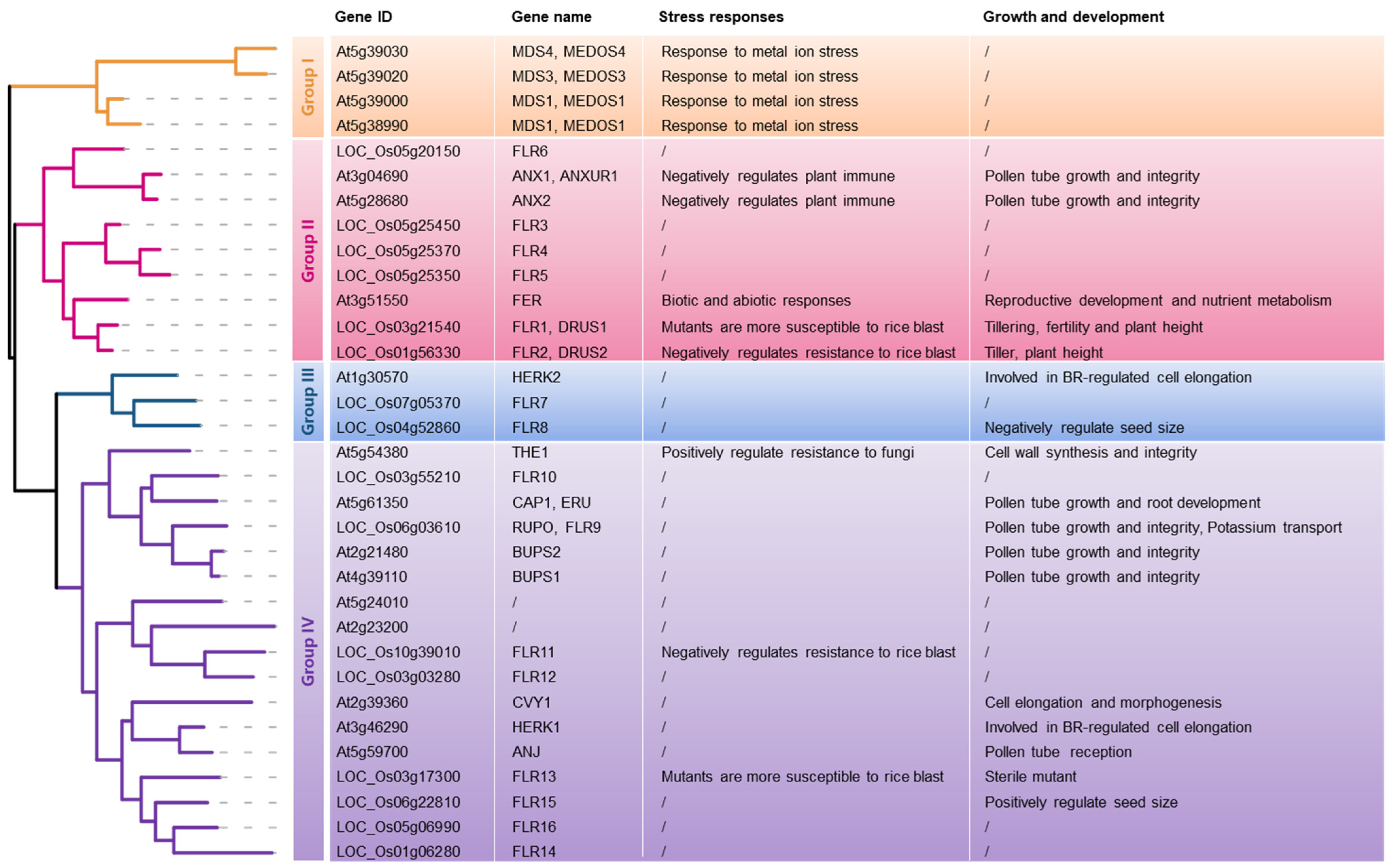
- Solis-Miranda, J.; Quinto, C. The CrRLK1L subfamily: One of the keys to versatility in plants. Plant Physiol. Biochem. 2021, 166, 88–102. [Google Scholar] [CrossRef]
- Solis-Miranda, J.; Fonseca-García, C.; Nava, N.; Pacheco, R.; Quinto, C. Genome-Wide Identification of the CrRLK1L Subfamily and Comparative Analysis of Its Role in the Legume-Rhizobia Symbiosis. Genes 2020, 11, 793. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Watson, J.M.; Stasnik, P.; Borowska, M.; Neuhold, J.; Berger, M.; Stolt-Bergner, P.; Schoft, V.; Hauser, M.T. Multiplex mutagenesis of four clustered CrRLK1L with CRISPR/Cas9 exposes their growth regulatory roles in response to metal ions. Sci. Rep. 2018, 8, 12182. [Google Scholar] [CrossRef] [PubMed]
- Mustamin, Y.; Akyol, T.Y.; Gordon, M.; Manggabarani, A.M.; Isomura, Y.; Kawamura, Y.; Bamba, M.; Williams, C.; Andersen, S.U.; Sato, S. FER and LecRK show haplotype-dependent cold-responsiveness and mediate freezing tolerance in Lotus japonicus. Plant Physiol. 2022, kiac533. [Google Scholar] [CrossRef]
- Schoenaers, S.; Balcerowicz, D.; Breen, G.; Hill, K.; Zdanio, M.; Mouille, G.; Holman, T.J.; Oh, J.; Wilson, M.H.; Nikonorova, N.; et al. The Auxin-Regulated CrRLK1L Kinase ERULUS Controls Cell Wall Composition during Root Hair Tip Growth. Curr. Biol. 2018, 28, 722–732.e6. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Trigo, S.; Blanco-Touriñán, N.; DeFalco, T.A.; Wells, E.S.; Gray, J.E.; Zipfel, C.; Smith, L.M. CrRLK1L receptor-like kinases HERK1 and ANJEA are female determinants of pollen tube reception. EMBO Rep. 2020, 21, e48466. [Google Scholar] [CrossRef]
- Pu, C.X.; Han, Y.F.; Zhu, S.; Song, F.Y.; Zhao, Y.; Wang, C.Y.; Zhang, Y.C.; Yang, Q.; Wang, J.; Bu, S.L.; et al. The Rice Receptor-Like Kinases DWARF AND RUNTISH SPIKELET1 and 2 Repress Cell Death and Affect Sugar Utilization during Reproductive Development. Plant Cell 2017, 29, 70–89. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Choi, J.-H.; Kim, J.-W.; Oh, M.-H. Identification of Feronia-interacting proteins in Arabidopsis thaliana. Genes Genom. 2022, 44, 1477–1485. [Google Scholar] [CrossRef]
- Malivert, A.; Erguvan, Ö.; Chevallier, A.; Dehem, A.; Friaud, R.; Liu, M.; Martin, M.; Peyraud, T.; Hamant, O.; Verger, S. FERONIA and microtubules independently contribute to mechanical integrity in the Arabidopsis shoot. PLoS Biol. 2021, 19, e3001454. [Google Scholar] [CrossRef]
- Haruta, M.; Gaddameedi, V.; Burch, H.; Fernandez, D.; Sussman, M.R. Comparison of the effects of a kinase-dead mutation of FERONIA on ovule fertilization and root growth of Arabidopsis. FEBS Lett. 2018, 592, 2395–2402. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Haruta, M.; Mours, D.S. Twenty Years of Progress in Physiological and Biochemical Investigation of RALF Peptides. Plant Physiol. 2020, 182, 1657–1666. [Google Scholar] [CrossRef]
- Vogler, H.; Santos-Fernandez, G.; Mecchia, M.A.; Grossniklaus, U. To preserve or to destroy, that is the question: The role of the cell wall integrity pathway in pollen tube growth. Curr. Opin. Plant Biol. 2019, 52, 131–139. [Google Scholar] [CrossRef]
- Somoza, S.C.; Sede, A.R.; Boccardo, N.A.; Muschietti, J.P. Keeping up with the RALFs: How these small peptides control pollen–pistil interactions in Arabidopsis. New Phytol. 2021, 229, 14–18. [Google Scholar] [CrossRef]
- Noble, J.A.; Seddon, A.; Uygun, S.; Bright, A.; Smith, S.E.; Shiu, S.H.; Palanivelu, R. The SEEL motif and members of the MYB-related REVEILLE transcription factor family are important for the expression of LORELEI in the synergid cells of the Arabidopsis female gametophyte. Plant Reprod. 2022, 35, 61–76. [Google Scholar] [CrossRef]
- Zhou, X.; Lu, J.; Zhang, Y.Q.; Guo, J.Z.; Lin, W.W.; Van Norman, J.M.; Qin, Y.; Zhu, X.Y.; Yang, Z.B. Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev. Cell 2021, 56, 1030–1042. [Google Scholar] [CrossRef]
- Dünser, K.; Gupta, S.B.; Herger, A.; Feraru, M.I.; Ringli, C.; Kleine-Vehn, J. Extracellular matrix sensing by FERONIA and Leucine-Rich Repeat Extensins controls vacuolar expansion during cellular elongation in Arabidopsis thaliana. EMBO J. 2019, 38, e100353. [Google Scholar] [CrossRef]
- Zhu, S.R.; Estévez, J.M.; Liao, H.D.; Zhu, Y.H.; Yang, T.; Li, C.Y.; Wang, Y.C.; Li, L.; Liu, X.M.; Pacheco, J.M.; et al. The RALF1-FERONIA Complex Phosphorylates eIF4E1 to Promote Protein Synthesis and Polar Root Hair Growth. Mol. Plant 2020, 13, 698–716. [Google Scholar] [CrossRef]
- Gronnier, J.; Franck, C.M.; Stegmann, M.; DeFalco, T.A.; Abarca, A.; von Arx, M.; Dünser, K.; Lin, W.W.; Yang, Z.B.; Kleine-Vehn, J.; et al. Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. eLife 2022, 11, e74162. [Google Scholar] [CrossRef]
- Gigli-Bisceglia, N.; van Zelm, E.; Huo, W.Y.; Lamers, J.; Testerink, C. Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 2022, 149, dev200363. [Google Scholar] [CrossRef]
- Shin, S.Y.; Park, J.-S.; Park, H.B.; Moon, K.B.; Kim, H.S.; Jeon, J.H.; Cho, H.S.; Lee, H.J. FERONIA Confers Resistance to Photooxidative Stress in Arabidopsis. Front. Plant Sci. 2021, 12, 714938. [Google Scholar] [CrossRef]
- Hansen, R.L.; Guo, H.Q.; Yin, Y.H.; Lee, Y.J. FERONIA mutation induces high levels of chloroplast-localized Arabidopsides which are involved in root growth. Plant J. 2019, 97, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Munns, R.; Burton, R.A.; Gilliham, M.; Wege, S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018, 269, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Z.; Zayed, O.; Yu, Z.P.; Jiang, W.; Zhu, P.P.; Hsu, C.C.; Zhang, L.R.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [PubMed]
- Herger, A.; Gupta, S.; Kadler, G.; Franck, C.M.; Boisson-Dernier, A.; Ringli, C. Overlapping functions and protein-protein interactions of LRR-extensins in Arabidopsis. PLoS Genet. 2020, 16, e1008847. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.-I. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, M.H.; Hong, W.J.; Moon, S.; Kim, S.T.; Park, S.K.; Jung, K.H. OsMTD2-mediated reactive oxygen species (ROS) balance is essential for intact pollen-tube elongation in rice. Plant J. 2021, 107, 1131–1147. [Google Scholar] [CrossRef]
- Wang, L.; Wang, D.D.; Yang, Z.H.; Jiang, S.; Qu, J.N.; He, W.; Liu, Z.M.; Xing, J.J.; Ma, Y.C.; Lin, Q.L.; et al. Roles of FERONIA-like receptor genes in regulating grain size and quality in rice. Sci. China Life Sci. 2021, 64, 294–310. [Google Scholar] [CrossRef]
- Yang, Z.H.; Xing, J.J.; Wang, L.; Liu, Y.; Qu, J.N.; Tan, Y.; Fu, X.Q.; Lin, Q.L.; Deng, H.F.; Yu, F. Mutations of two FERONIA-like receptor genes enhance rice blast resistance without growth penalty. J. Exp. Bot 2020, 71, 2112–2126. [Google Scholar] [CrossRef]
- Chen, Y.; Weckwerth, W. Mass spectrometry untangles plant membrane protein signaling networks. Trends Plant Sci. 2020, 25, 930–944. [Google Scholar] [CrossRef]
- Wang, Z.; Gou, X. Receptor-like protein kinases function upstream of MAPKs in regulating plant development. Int. J. Mol. Sci. 2020, 21, 7638. [Google Scholar] [CrossRef]
- Pandey, S. Plant receptor-like kinase signaling through heterotrimeric G-proteins. J. Exp. Bot. 2020, 71, 1742–1751. [Google Scholar] [CrossRef]
- Shumayla; Upadhyay, S. Shumayla; Upadhyay, S. An overview of receptor-like kinases in plants. 2023. In Plant Receptor-like Kinases; Academic Press: Cambridge, MA, USA, 2023. [Google Scholar]
- Luo, X.; Liu, J. Insights into receptor-like kinases-activated downstream events in plants. Sci. China Life Sci. 2018, 61, 1586–1588. [Google Scholar] [CrossRef]
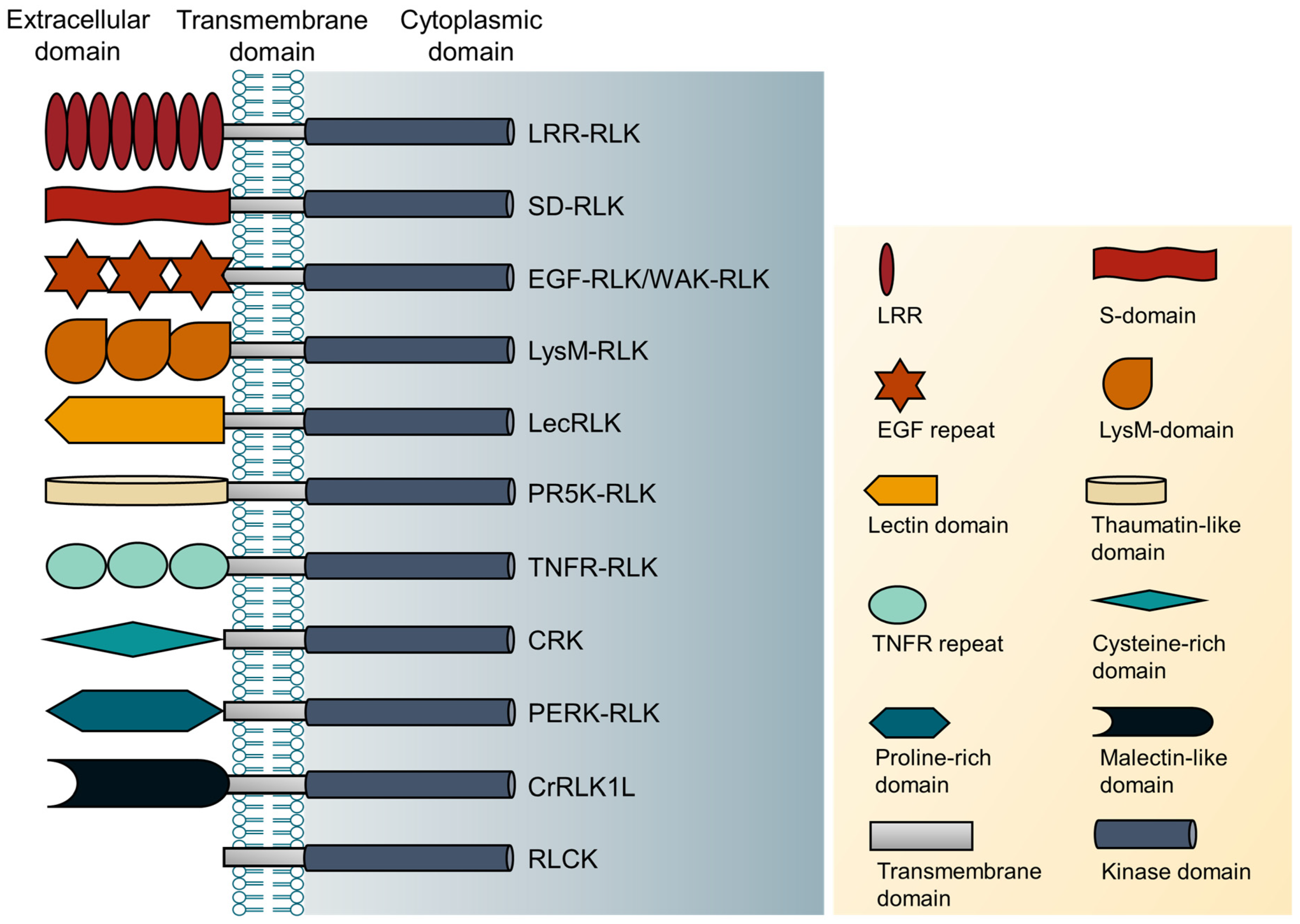
| No. | Type of RLKs | The Extracellular Domain of RLKs |
|---|---|---|
| 1. | Leucine-rich repeat receptor-like kinases (LRR-RLK) | Leucine-rich repeat domain |
| 2. | S-domain receptor-like kinases (SD-RLK) | S-domain |
| 3. | Epidermal growth factor-like kinases (EGF-RLK) | Epidermal growth factor repeat domain |
| 4. | Wall-associated receptor-like kinases (WAK-RLK) | EGF repeat domain- |
| 5. | Lysin motif-type receptor-like kinases (LysM-RLK) | LysM domain |
| 6. | Lectin receptor-like protein kinases (LecRLK) | Lectin domain |
| 7. | Pathogenesis related protein-5 like receptor kinases(PR5K-RLK) | Thaumatin-like domain |
| 8. | Tumor necrosis factor receptor-like protein kinases (TNFR-RLK) | Tumor necrosis factor receptor domain |
| 9. | Cysteine-rich receptor-like kinase (CRKs) | Cysteine-rich domain |
| 10. | Proline-rich extensin-like receptor kinases (PERK-RLK) | Proline-rich extensin-like domain |
| 11. | Catharanthus roseus receptor-like kinase 1-like (CrRLK1L) | Malectin-like domain |
| Plant Species | RLKs | Subfamily | Function | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana (Arabidopsis) | CLV1 | LRR-RLK | Meristem and flower development | [19,20,21] |
| ERfRLK | LRR-RLK | Cotyledon growth and ovule development | [25,26] | |
| BAM1/2 | LRR-RLK | Anther development | [21] | |
| RPK2 | LRR-RLK | Anther development | [21] | |
| AtVRLK1 | PR5K-RLK | Regulates secondary cell wall thickening; up-regulation of AtVRLK1 leads to defects in anther dehiscence | [27] | |
| AtPERK5/12 | PERK-RLK | Necessary for proper pollen tube growth | [28] | |
| BRI1 | LRR-RLK | Regulates cell elongation by mediation of BR signaling | [29,30,31,32,33] | |
| EMS1 | LRR-RLK | Anther development | [34] | |
| Oryza sativa (Rice) | OsERL | LRR-RLK | Anther lobe formation | [35] |
| TMS10/ TMS10L | LRR-RLK | Redundant control of male fertility under fluctuating temperatures; regulates tapetal degeneration and pollen development | [36] | |
| OsLecRK5 | LecRLK | Regulates callose biosynthesis during pollen development | [37] | |
| OsLSK1 | SD-RLK | Overexpression of OsLSK1 extracellular domain improves panicle architecture and grain yield | [38] | |
| OsER1 | LRR-RLK | Negative regulator of spikelet number per panicle | [39] | |
| SERK2 | LRR-RLK | Overexpression of SERK2 enhances grain size and salt resistance | [40] | |
| OsRPK1 | LRR-RLK | Negatively regulates root development | [41] | |
| OsESG1 | SD-RLK | Regulates early crown root development and drought resistance | [42] | |
| Triticum aestivum L. (Wheat) | TaBRI1 | LRR-RLK | Early flowering and seed yield enhancement in Arabidopsis | [43,44,45] |
| Plant Species | RLKs | Subfamily | Biotic Stress Type/Name | Function | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana (Arabidopsis) | CDG1 | RLCK | Bacterial disease/Pseudomonas syringae | Negatively regulates Arabidopsis pattern-triggered immunity (PTI) | [68] |
| AtSERK1/AtSERK2 | LRR-RLK | Bacterial disease/Pseudomonas syringae | Resistance to bacterial leaf blight and fungal infection | [69] | |
| CRK28/ CRK29 | CRK | Bacterial disease/P. syringae | Enhances plant immune responses | [70] | |
| AtBAK1 | LRR-RLK | Fungal diseases/ Cladosporium fulvum | Triggers immune signaling to promote plant resistance against pathogens | [71] | |
| BAM1 | LRR-RLK | Viral disease/Tobacco mosaic virus (TMV) | Involvement in the early stages of TMV spread and cell-to-cell movement | [72] | |
| NIK1 | LRR-RLK | Viral disease/begomovirus Cabbage leaf curl virus; Bacterial disease /P. syringae DC3000 and ES4326 | Positively regulates plant antiviral immunity; Negatively regulates plant of antibacterial immunity | [73] | |
| Oryza sativa (Rice) | rrsRLK | RLCK | Bacterial disease/Xanthomonas oryzae pv. oryzae (Xoo) | Δrrsrlk resistant to bacterial leaf blight in Rice | [74] |
| Pi65 | LRR-RLK | Fungal diseases/Magnaporthe oryzae | Overexpression of Pi65 enhanced rice blast resistance | [75] | |
| SDS2 | SD-RLK | Fungal diseases/M. oryzae | SDS2 overexpression enhanced resistance to M. Oryzae | [76] | |
| OsSOBIR1 | LRR-RLK | Viral disease/Rice black-streaked dwarf virus(RBSDV) | Regulates the PTI response and rice antiviral defense to RBSDV | [77] | |
| OsLRR-RLK1 | LRR-RLK | Herbivore attack/striped stem borer (SSB) Chilo suppressalis | Against the chewing herbivore SSB | [78] | |
| OsLRR-RLK2 | LRR-RLK | Herbivore attack/brown planthopper (BPH, Nilaparvata lugens) | Negatively regulates the resistance of rice to BPH | [79] | |
| Triticum aestivum L. (Wheat) | TaXa21 | LRR-RLK | Fungal diseases/Puccinia striiformis f. sp. tritici | Positive regulator of wheat High-temperature seedling-plant resistance to P. Striiformis f. Sp. Tritici | [80] |
| TaCRK10 | CRK | Fungal diseases/P. striiformis f. sp. tritici | High-temperature seedling-plant resistance to stripe rust caused by fungal pathogen P. striiformis f. Sp. Tritici | [81] |
| Plant Species | RLKs | Subfamily | Biotic Stress Type/Name | Function | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana (Arabidopsis) | RLK7 | LRR-RLK | Drought stress | Regulates immune responses and stomatal closure | [90,91] |
| RPK1/BAK1 | LRR-RLK | Drought stress | Positively regulates ABA-induced stomatal closure | [92] | |
| Oryza sativa (Rice) | LP2 | LRR-RLK | Drought stress | Negative regulator in drought response | [93] |
| HSL3 | LRR-RLK | Drought stress | Regulates stomatal closure and drought stress response | [94] | |
| OsSIT1 | LecRLK | Salt stress | Negatively regulates salt sensitivity | [95] | |
| OsSTLK | LRR-RLK | Salt stress | Positive regulator of salt stress tolerance | [96] | |
| OsSTRK1 | RLCK | Salt stress | Positively regulates salt and oxidative stress tolerance | [97] | |
| OsWAK11 | WAK-RLK | Metal stress/aluminum and copper | Regulates resistance to aluminum and copper | [98] | |
| LRK10-L | PR5K-RLK | Metal stress/cadmium | Regulates chromium stress | [99] | |
| DUF26 | CRK | Metal stress/cadmium | Regulates chromium stress | [99] | |
| Glycine soja (Soybean) | GsLRPK | LRR-RLK | Cold stress | Positive regulator to cold stress tolerance | [100] |
| Medicago truncatula | MtCTLK1 | LRR-RLK | Cold stress | Positive regulates cold tolerance | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Q.; Feng, Y.; Xue, J.; Chen, P.; Zhang, A.; Yu, Y. Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses. Plants 2023, 12, 427. https://doi.org/10.3390/plants12030427
Zhu Q, Feng Y, Xue J, Chen P, Zhang A, Yu Y. Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses. Plants. 2023; 12(3):427. https://doi.org/10.3390/plants12030427
Chicago/Turabian StyleZhu, Qingfeng, Yanzhao Feng, Jiao Xue, Pei Chen, Aixia Zhang, and Yang Yu. 2023. "Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses" Plants 12, no. 3: 427. https://doi.org/10.3390/plants12030427
APA StyleZhu, Q., Feng, Y., Xue, J., Chen, P., Zhang, A., & Yu, Y. (2023). Advances in Receptor-like Protein Kinases in Balancing Plant Growth and Stress Responses. Plants, 12(3), 427. https://doi.org/10.3390/plants12030427






