Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance
Abstract
:1. Introduction
2. Influence of Cold Stress on the Growth and Development of Soybeans
3. Candidate Genes/QTLs Associated with Cold Tolerance in Soybean
| Candidate Genes/QTLs/Associated Markers | Chromosome/Linkage Group | References |
|---|---|---|
| APXI | 11(B1) | [30] |
| T | 6(C2) | [31,32,36,37,38] |
| E1 | 6(C2) | [32,38] |
| E4 | 20(I) | [31,38] |
| Ln | 20(I) | [33] |
| PI | 9(K) | [33] |
| Dt1 | 19(L) | [33] |
| E3 | 19(L) | [31,38] |
| qCTTSW1 | 6(C2) | [31] |
| qCTTSW2 | 19(L) | [31] |
| qCTTSW3 | 12(H) | [31] |
| GmIRCHS | 8(A2) | [35] |
| Sat_342 | 14(B2) | [35] |
| qCS8-1 | 8(A2) | [39] |
| qCS11-1 | 8(B1) | [39] |
| Ic | 8(A2) | [35,40] |
| S07_42231812 | 7(M) | [41] |
| S11_30574868 | 11(B1) | [41] |
| S13_33041524 | 13(F) | [41] |
| Sat_271 | 5(A1) | [8] |
| Satt225 | 5(A1) | |
| Sat_331 | 11(B1) | |
| Satt168 | 14(B2) | |
| Satt577 | 14(B2) | |
| Satt338 | 4(C1) | |
| Satt640 | 6(C2) | |
| Satt041 | 1(D1b) | |
| Satt271 | 1(D1b) | |
| Satt458 | 17(D2) | |
| Satt669 | 17(D2) | |
| Satt651 | 15(E) | |
| Satt142 | 12(H) | |
| Satt253 | 12(H) | |
| Satt353 | 12(H) | |
| Satt440 | 20(I) | |
| Satt249 | 16(J) | |
| Sat_126 | 9(K) | |
| Satt240 | 9(K) | |
| Satt349 | 9(K) | |
| Satt513 | 19(L) | |
| Sat_244 | 7(M) | |
| Satt323 | 7(M) | |
| Satt336 | 7(M) | |
| Satt540 | 7(M) | |
| Sat_192 | 1(D1b) | |
| Satt663 | 13(F) | |
| Sat_020 | 9(K) |
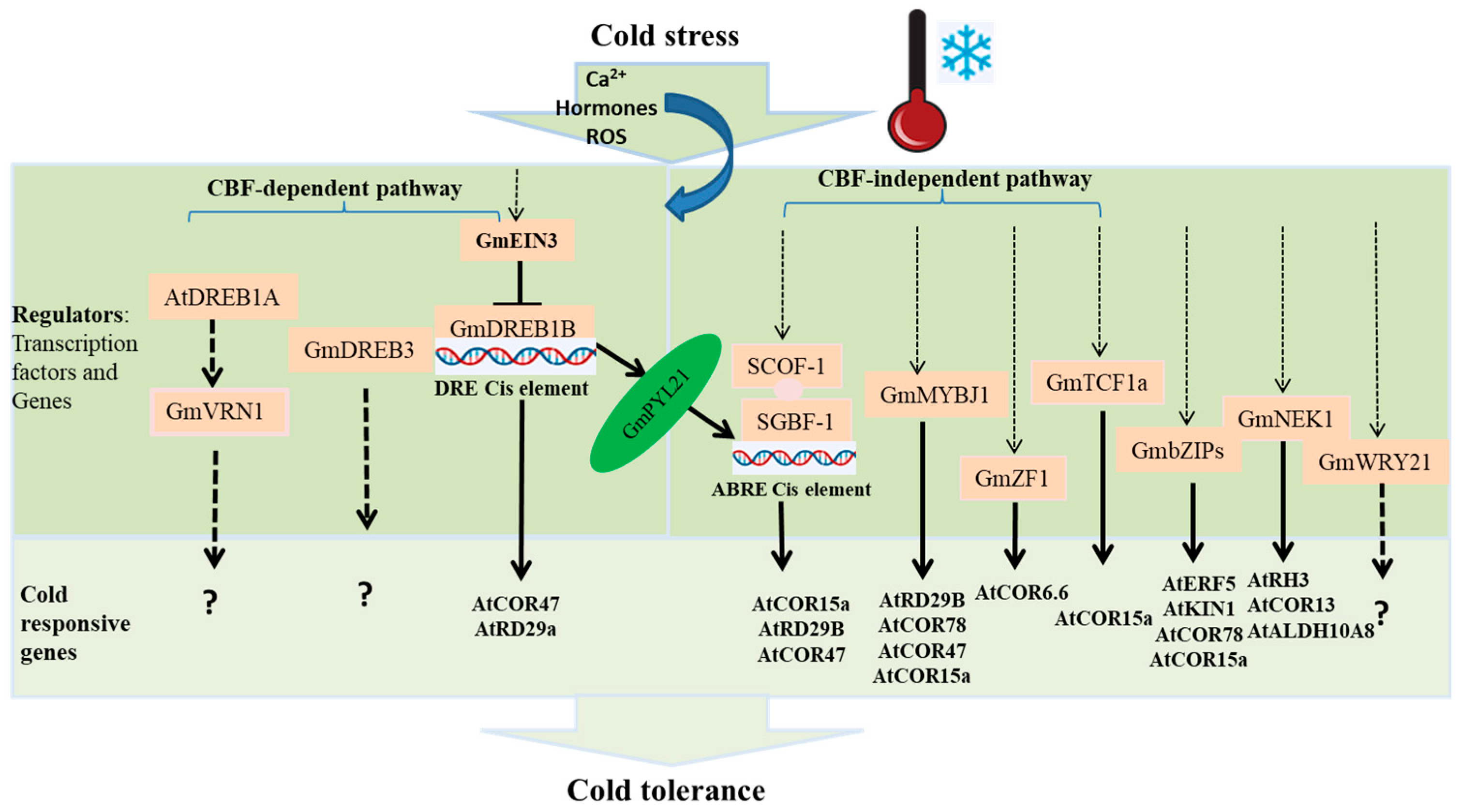
4. Soybean Cold Responsive Regulators and Associated Cold Regulated (COR) Genes
4.1. Soybean CBF-Dependent Cold Response Regulatory Pathway
4.2. Other Soybean Cold Response Regulatory Pathways
| Factor | Gene Characteristics | Effect on Cold Tolerance | References |
|---|---|---|---|
| GmDREB1s | DREB genes | Arabidopsis and soybean, Positive | [48] |
| GmDREB3 | DREB gene | Arabidopsis, Positive | [46,47] |
| GmNEK1 | NIMA-related kinase gene | Arabidopsis, Positive | [53] |
| GmEIN3 | Ethylene insensitive gene | Soybean, Negative | [51] |
| GmMYBJ1 | MYB transcription factor | Arabidopsis, Positive | [62] |
| SCOF-1 | C2H2 zinc finger gene | Arabidopsis and tabacco, Positive | [50] |
| GmZF1 | C2H2 zinc finger gene | Arabidopsis, Positive | [67] |
| GmPYL21 | ABA receptor family gene | Soybean, Positive | [48] |
| Glyma11g13220(GmVRN1-like) | Vernalization pathway gene | Arabidopsis, Positive | [59] |
| GmTCF1a | RCC1 family gene | Arabidopsis, Positive | [68] |
| GmWRKY21 | WRKY-type transcription factor | Arabidopsis, Positive | [63] |
| GmbZIP44, GmbZIP62 and GmbZIP78 | bZIP-transcription factor | Arabidopsis, Positive | [64] |
| Hsp 70 | Heat shock protein genes | Soybean, Positive | [72] |
5. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, K. (Ed.) Chemistry and Nutritional Value of Soybean. In Soybeans: Chemistry, Technology, and Utilization; Springer: Boston, MA, USA, 1997; p. 532. [Google Scholar] [CrossRef]
- Hungria, M.; Franchini, J.C.; Campo, R.J.; Graham, P.H. The Importance of Nitrogen Fixation to Soybean Cropping in South America. In Nitrogen Fixation in Agriculture, Forestry, Ecology, and the Environment; Werner, D., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 25–42. [Google Scholar] [CrossRef]
- Jia, H.; Jiang, B.; Wu, C.; Lu, W.; Hou, W.; Sun, S.; Yan, H.; Han, T. Maturity Group Classification and Maturity Locus Genotyping of Early-Maturing Soybean Varieties from High-Latitude Cold Regions. PLoS ONE 2014, 9, e94139. [Google Scholar] [CrossRef] [PubMed]
- Lu, W. The soybean cultivation technique in northern area of Heilongjiang province. China Seed Ind. 2011, 33, 59–60. [Google Scholar]
- Nedoluzhko, A.V.; Dorokhov, D.B. Study of the biosafety of genetically modified soybean in the center of its origin and diversity in the Far East of the Russian Federation. Cytol. Genet. 2007, 41, 190–198. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Li, K.; Zou, Y.; Chen, L.; Li, X. Identification of cold-responsive miRNAs and their target genes in nitrogen-fixing nodules of soybean. Int. J. Mol. Sci. 2014, 15, 13596–13614. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Browse, J. Cold comfort farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 2000, 23, 893–902. [Google Scholar] [CrossRef]
- Zhang, W.B.; Jiang, H.W.; Qiu, P.C.; Liu, C.Y.; Chen, F.L.; Xin, D.W.; Li, C.D.; Hu, G.H.; Chen, Q.S. Genetic overlap of QTL associated with low-temperature tolerance at germination and seedling stage using BILs in soybean. Can. J. Plant Sci. 2012, 92, 1381–1388. [Google Scholar] [CrossRef] [Green Version]
- Ruelland, E.; Vaultier, M.-N.; Zachowski, A.; Hurry, V. Cold Signalling and Cold Acclimation in Plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Pearce, R.S. Plant freezing and damage. Ann. Bot. 2001, 87, 417–424. [Google Scholar] [CrossRef]
- Steponkus, P.L. Role of the plasma membrane in freezing injury and cold acclimation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1984, 35, 543–584. [Google Scholar] [CrossRef]
- Takahashi, R.; Shimosaka, E. cDNA sequence analysis and expression of two cold-regulated genes in soybean. Plant Sci. 1997, 123, 93–104. [Google Scholar] [CrossRef]
- McKersie, B.D.; Leshem, Y.Y. Chilling Stress. Stress and Stress Coping in Cultivated Plants; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Alsajri, F.A.; Singh, B.; Wijewardana, C.; Irby, J.T.; Gao, W.; Reddy, K.R. Evaluating Soybean Cultivars for Low- and High-Temperature Tolerance during the Seedling Growth Stage. Agronomy 2019, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- Duke, S.H.; Schrader, L.E.; Miller, M.G.; Niece, R.L. Low Temperature Effects on Soybean (Glycine max [L.] Merr. cv. Wells) Free Amino Acid Pools during Germination. Plant Physiol. 1978, 62, 642–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staniak, M.; Kocira, A.; Czopek, K.; Stepien-Warda, A.; Orybys, M. Cold Stress during Flowering Alters Plant Structure, Yield and Seed Quality of Different Soybean Genotypes. Agronomy 2021, 11, 2059. [Google Scholar] [CrossRef]
- Liu, X.; Jin, J.; Wang, G.; Herbert, S.J. Soybean yield physiology and development of high-yielding practices in Northeast China. Field Crops Res. 2018, 105, 157–171. [Google Scholar] [CrossRef]
- William, J.; Bramlage, A.; Leopold, A.C.; Parrish, D.J. Chilling Stress to Soybeans during Imbibition. Plant Physiol. 1978, 61, 525–529. [Google Scholar]
- Camara GM, S.; Sediyama, T.; Dourado-Neto, D.; Bernardes, M.S. Influência do fotoperíodo e da temperatura do ar no crescimento, floração e maturação da soja (Glycine max (L.) merrill). Sci. Agric. 1997, 54, 149–154. [Google Scholar] [CrossRef]
- Kurosaki, H.; Yumoto, S. Effects of low temperature and shading during flowering on the yield components in soybeans. Plant Prod. Sci. 2003, 6, 17–23. [Google Scholar] [CrossRef]
- Funatsuki, H.; Ohnishi, S. Recent Advances in Physiological and Genetic Studies on Chilling Tolerance in Soybean. Jpn. Agric. Res. Q. 2009, 43, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, S.; Miyoshi, T.; Shirai, S. Low temperature stress at different flower developmental stages affects pollen development, pollination, and pod set in soybean. Environ. Exp. Bot. 2010, 69, 56–62. [Google Scholar] [CrossRef]
- Gass, T.; Schori, A.; Fossati, A.; Soldati, A.; Stamp, P. Cold tolerance of soybean (Glycine max (L.) Merr.) during the reproductive phase. Eur. J. Agron. 1996, 5, 71–88. [Google Scholar] [CrossRef]
- Hume, D.J.; Jackson AK, H. Frost tolerance in soybeans. Crop Sci. 1981, 21, 689–692. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muncan, J.; Kuroki, S.; Tsenkova, R. Aquaphotomics Research of Cold Stress in Soybean Cultivars with Different Stress Tolerance Ability: Early Detection of Cold. Molecules 2022, 27, 744. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fang, Y.; Li, X.; Zeng, K.; Chen, H.; Zhang, H.; Yang, H. Identification of soybean drought—Tolerant genotypes and loci correlated with agronomic traits contributes new candidate genes for breeding. Plant Mol. Biol. 2019, 102, 109–122. [Google Scholar] [CrossRef]
- Takahashi, R.; Benitez, E.; Funatsuki, H.; Ohnishi, S. Soybean maturity and pubescence color genes improve chilling tolerance. Crop Sci. 2005, 45, 1387–1393. [Google Scholar] [CrossRef]
- Funatsuki, H.; Kurosaki, H.; Murakami, T.; Matsuba, S.; Kawaguchi, K.; Yumoto, S.; Sato, Y. Deficiency of a cytosolic ascorbate peroxidase associated with chilling tolerance in soybean. Theor. Appl. Genet. 2003, 106, 494–502. [Google Scholar] [CrossRef]
- Funatsuki, H.; Kawaguchi, K.; Matsuba, S.; Sato, Y.; Ishimoto, M. Mapping of QTL associated with chilling tolerance during reproductive growth in soybean. Theor. Appl. Genet. 2005, 111, 851–861. [Google Scholar] [CrossRef]
- Kurosaki, H.; Yumoto, S.; Matsukawa, I. Correlation of cold-weather tolerance with pubescence color and flowering time in yellow hilum soybeans in Hokkaido. Breed. Sci. 2004, 54, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Palmer, R.G.; Pfeiffer, T.W.; Buss, G.R.; Kilen, T.C. Qualitative genetics. In Soybeans: Improvement, Production, and Uses, 3rd ed.; Shibles, R., Harper, J., Wilson, R., Shoemaker, R., Eds.; ASA; CSSA; SSSA: Madison, WI, USA, 2004; pp. 137–233. [Google Scholar]
- Githiri, S.M.; Yang, D.; Khan, N.A.; Xu, D.; Komatsuda, T.; Takahashi, R. QTL Analysis of Low Temperature—Induced Browning in Soybean Seed Coats. J. Hered. 2007, 98, 360–366. [Google Scholar] [CrossRef]
- Ohnishi, S.; Funatsuki, H.; Kasai, A.; Kurauchi, T.; Yamaguchi, N.; Takeuchi, T.; Yamazaki, H.; Kurosaki, H.; Shirai, S.; Miyoshi, T.; et al. Variation of GmIRCHS (Glycine max inverted-repeat CHS pseudogene) is related to tolerance of low temperature-induced seed coat discoloration in yellow soybean. Theor. Appl. Genet. 2011, 122, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.J.; Voldeng, H.D.; Guillemette, R.J.; Cober, E.R. Yield of cool-season soybean lines differing in pubescence color and density. Agron. J. 1997, 89, 218–221. [Google Scholar] [CrossRef]
- Takahashi, R.; Abe, J. Genetic and linkage analysis of low temperature-induced browning in soybean seed coats. J. Hered. 1994, 85, 447–450. [Google Scholar] [CrossRef]
- Takahashi, R.; Abe, J. Soybean maturity genes associated with seed coat pigmentation and cracking in re-sponse to low temperatures. Crop Sci. 1999, 39, 1657–1662. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Taguchi-shiobara, F.; Sayama, T. Quantitative Trait Loci associated with Tolerance to Seed Cracking under Chilling Temperatures in Soybean. Crop Sci. 2015, 55, 2100–2107. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Hagihara, S.; Hirai, D. Field assessment of a major QTL associated with tolerance to cold-induced seed coat discoloration in soybean. Breed. Sci. 2019, 69, 521–528. [Google Scholar] [CrossRef] [Green Version]
- Jähne, F.; Balko, C.; Hahn, V.; Würschum, T. Cold stress tolerance of soybeans during flowering: QTL mapping and efficient selection strategies under controlled conditions. Plant Breed. 2019, 138, 708–720. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Knight, M.R.; Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012, 195, 737–751. [Google Scholar] [CrossRef]
- Huber, A.; Bauerle, T. Long-distance plant signaling pathways in response to multiple stressors: The gap in knowledge. J. Exp Bot. 2016, 76, 2063–2079. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, X.-M.; Lin, H.-X.; Kang, C. Crop Improvement through Temperature Resilience. Annu. Rev. Plant Biol. 2019, 70, 753–780. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 2009, 60, 121–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasreen, S.; Amudha, J.; Pandey, S.S. Isolation and characterization of Soybean DREB 3 transcriptional activator. J. Appl. Biol. Biotechnol. 2013, 1, 9–12. [Google Scholar] [CrossRef]
- Kidokoro, S.; Watanabe, K.; Ohori, T.; Moriwaki, T.; Maruyama, K.; Mizoi, J.; Myint Phyu Sin Htwe, N.; Fujita, Y.; Sekita, S.; Shinozaki, K.; et al. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015, 81, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Randall, S.K. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 2016, 246, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.C.; Lee, S.H.; Cheong, Y.H.; Yoo, C.M.; Lee, S.I.; Chun, H.J.; Yun, D.J.; Hong, J.C.; Lee, S.Y.; Lim, C.O.; et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001, 25, 247–259. [Google Scholar] [CrossRef]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K.; Randall, S.K. The Ethylene Signaling Pathway Negatively Impacts CBF / DREB-Regulated Cold Response in Soybean (Glycine max). Front. Plant Sci. 2019, 10, 121. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, Y.; Koehler, G.; Blacklock, B.J.; Randall, S.K. Dehydrin expression in soybean. Plant Physiol. Biochem. 2013, 70, 213–220. [Google Scholar] [CrossRef]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Shen, M.; Ma, J.B.; Zhang, W.K.; Lin, Q.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean NIMA-Related Kinase1 Promotes Plant Growth and Improves Salt and Cold Tolerance. Plant Cell Physiol. 2017, 58, 1268–1278. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, M.; Jaiswal, A.; Taj, G.; Jaiswal, J.P.; Qureshi, M.I.; Singh, N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012, 91, 385–395. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomaschow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta—Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Guo, L.; Cui, F.; Shen, Y.; Ye, X.; Deng, D.; Wang, S.; Zhu, J.; Wu, W. Innovations and stepwise evolution of CBFs/DREB1s and their regulatory networks in angiosperms. J. Integr. Plant Biol. 2022, 64, 2111–2125. [Google Scholar] [CrossRef] [PubMed]
- Suo, H.; Lü, J.; Ma, Q.; Yang, C.; Zhang, X.; Meng, X.; Huang, S.; Nian, H. The AtDREB1A transcription factor up-regulates expression of a vernalization pathway gene, GmVRN1-like, delaying flowering in soybean. Acta Physiol. Plant. 2016, 38, 137. [Google Scholar] [CrossRef] [Green Version]
- Lü, J.; Suo, H.; Yi, R.; Ma, Q.; Nian, H. Glyma11g13220, a homolog of the vernalization pathway gene VERNALIZATION 1 from soybean [Glycine max (L.) Merr.], promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 2015, 15, 232. [Google Scholar] [CrossRef] [Green Version]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef]
- Su, L.T.; Li, J.W.; Liu, D.Q.; Zhai, Y.; Zhang, H.J.; Li, X.W.; Zhang, Q.L.; Wang, Y.; Wang, Q.Y. A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 2014, 538, 46–55. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Liao, Y.; Wei, H.Z.; Hao, W.Y.; Jian, A.T.; Liu, H.Y.; Chen JZ, S. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Ishitani, M.; Xiong, L.; Stevenson, B.; Zhul, J. Genetic Analysis of Osmotic and Cold Stress Signal Transduction in Arabidopsis: Lnteractions and Convergence of Abscisic Acid-Dependent and Abscisic Acid-lndependent Pathways. Plant Cell 1997, 9, 1935–1949. [Google Scholar] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H. Interaction between two cis -acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Jiang, L.; Ma, X.; Xu, Z.; Liu, M.; Shan, S. A Soybean C2H2-Type Zinc Finger Gene GmZF1 Enhanced Cold Tolerance in Transgenic Arabidopsis. PLoS ONE 2014, 9, e109399. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, Y.; Cloix, C.; Li, K.; Jenkins, G.I. The Arabidopsis RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef]
- Kodaira, K.S.; Qin, F.; Tran, L.S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 2011, 157, 742–756. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Caban, M.; Calvet, P.; Vincens, P.; Boudet, A.M. Characterization of chilling-acclimation-related proteins in soybean and identification of one as a member of the heat shock protein (HSP 70) family. Planta 1993, 190, 346–353. [Google Scholar] [CrossRef]
- Tyczewska, A.; Gracz, J.; Kuczyński, J.; Twardowski, T. Deciphering the soybean molecular stress response via high-throughput approaches. Acta ABP Biochim. Pol. 2016, 63, 631–643. [Google Scholar]
- Xu, S.; Liu, N.; Mao, W.; Hu, Q.; Wang, G.; Gong, Y. Identification of chilling-responsive microRNAs and their targets in vegetable soybean (Glycine max L.). Sci. Rep. 2016, 6, 26619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczyński, J.; Twardowski, T.; Nawracała, J.; Tyczewska, J.G.A. Chilling stress tolerance of two soya bean cultivars: Phenotypic and molecular responses. J. Agron. Crop Sci. 2020, 206, 759–772. [Google Scholar] [CrossRef]
- Wang, X.; Chang, X.; Jing, Y.; Zhao, J.; Fang, Q.; Sun, M.; Zhang, Y.; Li, W.; Li, Y. Identification and functional prediction of soybean CircRNAs involved in low-temperature responses. J. Plant Physiol. 2020, 250, 153188. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.; Baiya, S.; Lai, Z. An advanced systems biology framework of feature engineering for cold tolerance genes discovery from integrated omics and non-omics data in soybean. Front. Plant Sci. 2022, 13, 1019709. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, N.; Li, S.; Liao, W.; Shen, J.; Peng, M. The regulatory effects of MeTCP4 on cold stress tolerance in Arabidopsis thaliana: A transcriptome analysis. Plant Physiol. Biochem. 2019, 138, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 1, 1690–1704. [Google Scholar] [CrossRef] [Green Version]
- Bhat, K.A.; Masi, A. Low Temperature Stress Tolerance: An Insight into the Omics Approaches for Legume Crops. Front. Plant Sci. 2022, 13, 888710. [Google Scholar] [CrossRef]
- Buti, M.; Pasquariello, M.; Ronga, D.; Milc, J.A.; Pecchioni, N.; Pucciariello, C.; Perata, P.; Francia, E. Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp. Japonica seedlings. Funct. Integr. Genom. 2018, 18, 627–644. [Google Scholar] [CrossRef]
- Guan, S.; Xu, Q.; Ma, D.; Zhang, W.; Xu, Z.; Zhao, M.; Guo, Z. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 2019, 685, 96–105. [Google Scholar] [CrossRef]
- Cheng, L.; Gao, X.; Li, S.; Shi, M.; Javeed, H.; Jing, X.; Yang, G.; He, G. Proteomic analysis of soybean [Glycine max (L.) Meer.] seeds during imbibition at chilling temperature. Mol. Breed. 2010, 26, 1–17. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsegaw, M.; Zegeye, W.A.; Jiang, B.; Sun, S.; Yuan, S.; Han, T.; Wu, T. Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants 2023, 12, 459. https://doi.org/10.3390/plants12030459
Tsegaw M, Zegeye WA, Jiang B, Sun S, Yuan S, Han T, Wu T. Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants. 2023; 12(3):459. https://doi.org/10.3390/plants12030459
Chicago/Turabian StyleTsegaw, Mesfin, Workie Anley Zegeye, Bingjun Jiang, Shi Sun, Shan Yuan, Tianfu Han, and Tingting Wu. 2023. "Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance" Plants 12, no. 3: 459. https://doi.org/10.3390/plants12030459
APA StyleTsegaw, M., Zegeye, W. A., Jiang, B., Sun, S., Yuan, S., Han, T., & Wu, T. (2023). Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants, 12(3), 459. https://doi.org/10.3390/plants12030459








