Accelerated Breeding for Helianthus annuus (Sunflower) through Doubled Haploidy: An Insight on Past and Future Prospects in the Era of Genome Editing
Abstract
:1. Introduction
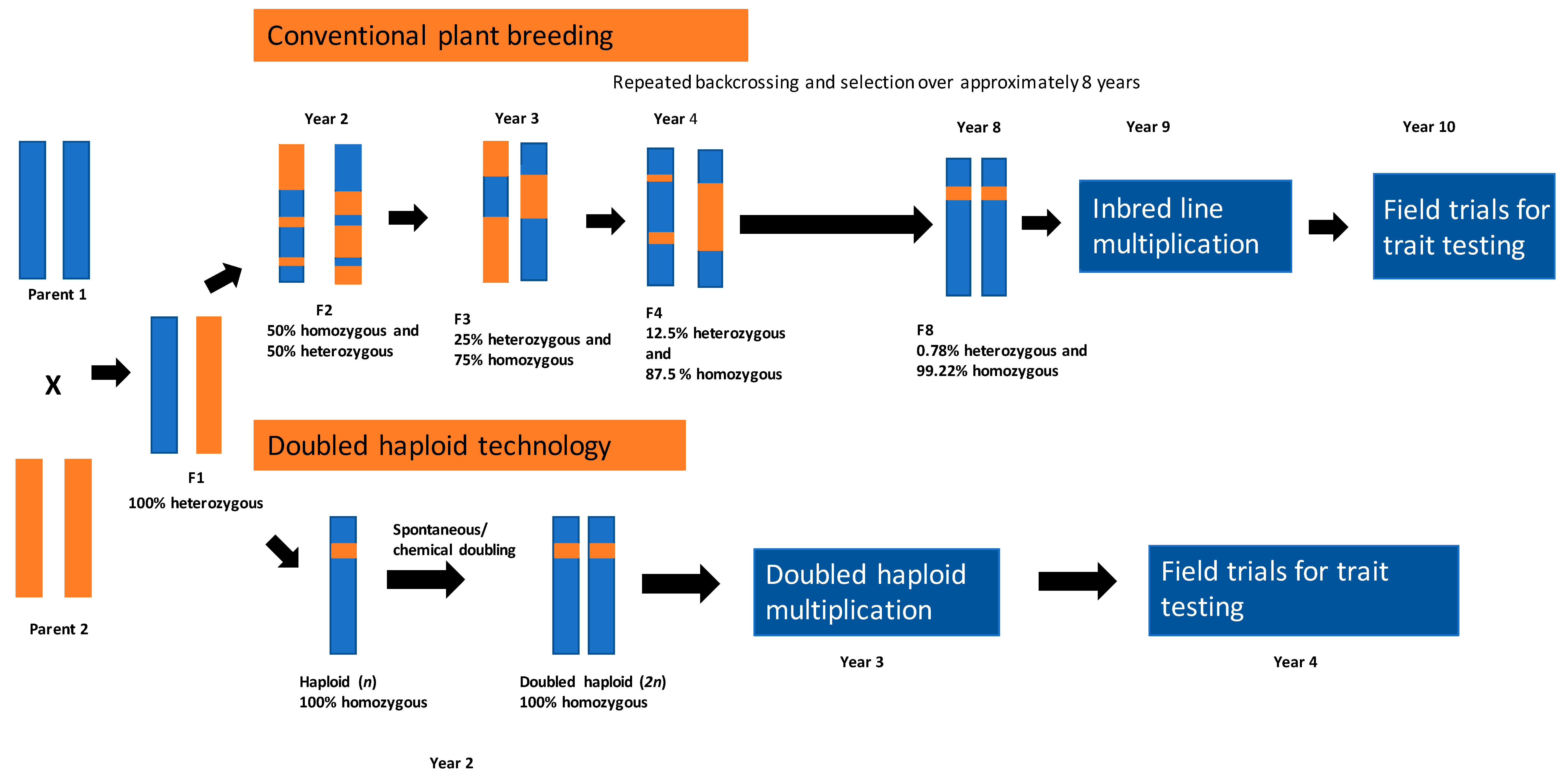
2. Doubled Haploids: Induction Methods and Their Role in Plant Breeding and Crop Improvement
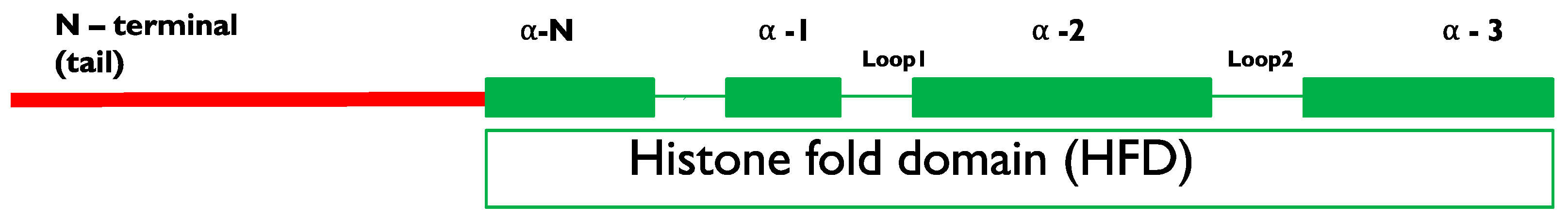
Haploid Induction by Meiotic/Mitotic Process Manipulations e.g., Centromeric Histone Protein 3 (CenH3) Modification
3. Methods for CenH3 Modification
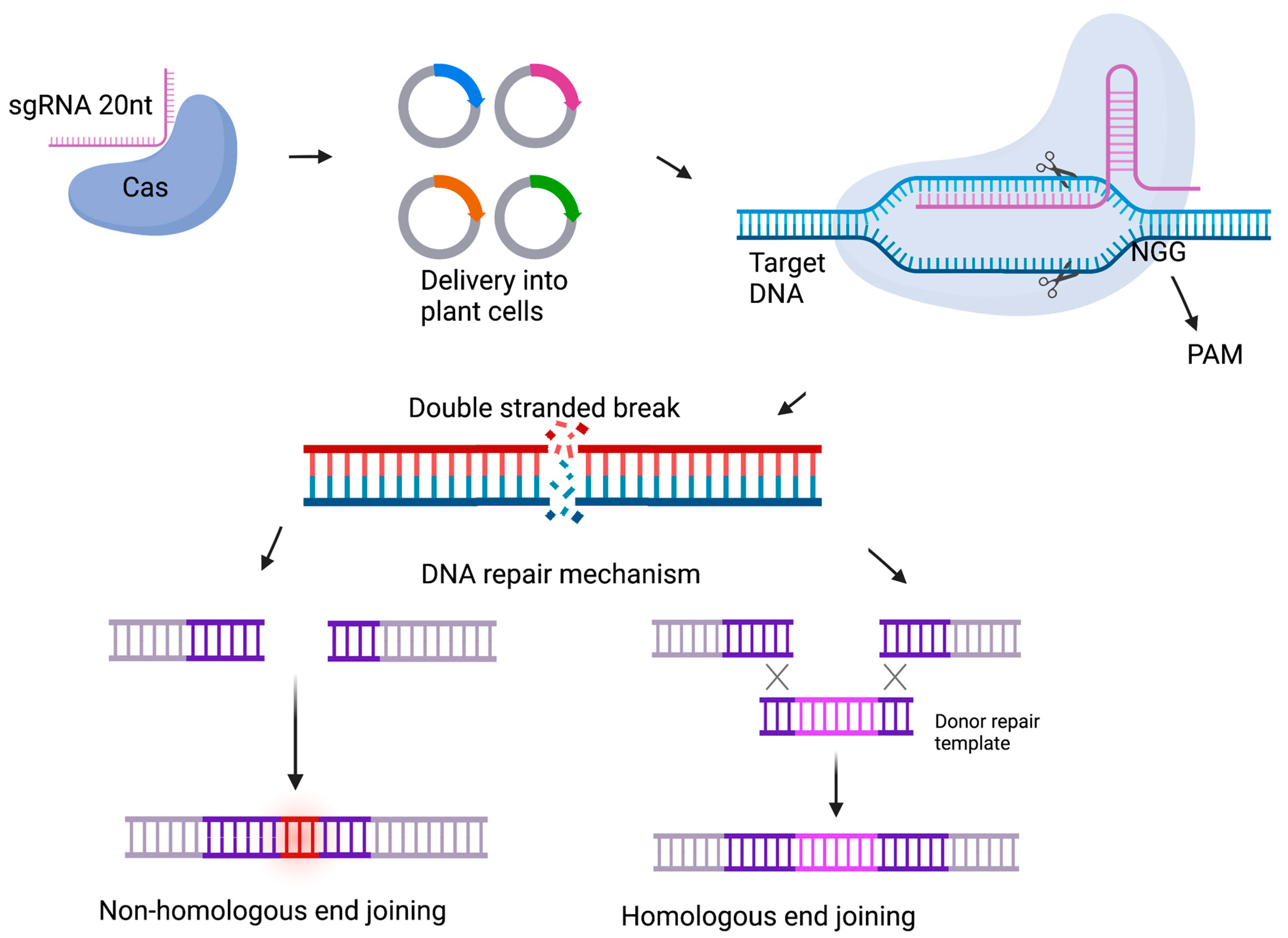
4. Delivery Methods for Genome Editing Components
4.1. Agrobacterium Tumefaciens-Mediated Transformation
4.2. Particle Bombardment
4.3. Viral Vectors for Plant Genome Editing
5. Prospects for Doubled Haploids in Sunflower
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pilorgé, E. Sunflower in the Global Vegetable Oil System: Situation, Specificities and Perspectives. Oilseeds Fats Crop. Lipids 2020, 27, 37. [Google Scholar] [CrossRef]
- Feng, J.; Jan, C.-C.; Seiler, G. Breeding, Production, and Supply Chain of Confection Sunflower in China. OCL 2022, 29, 11. [Google Scholar] [CrossRef]
- Davey, M.R.; Jan, M. Sunflower (Helianthus Annuus L.): Genetic Improvement Using Conventional and In Vitro Technologies. J. Crop Improv. 2010, 24, 349–391. [Google Scholar] [CrossRef]
- Blinkov, A.O.; Varlamova, N.V.; Kurenina, L.V.; Khaliluev, M.R. The Production of Helianthus Haploids: A Review of Its Current Status and Future Prospects. Plants 2022, 11, 2919. [Google Scholar] [CrossRef] [PubMed]
- Drumeva, M.; Yankov, P.; Nenova, N.; Shindrova, P. Investigation on the Resistance of Doubled Haploid Sunflower Lines to Some Biotic Factors. Genet. Breed. 2014, 6, 11–13. [Google Scholar]
- Kallamadi, P.R.; Mulpuri, S. Ploidy Analysis of Helianthus Species by Flow Cytometry and Its Use in Hybridity Confirmation. Nucleus 2016, 59, 123–130. [Google Scholar] [CrossRef]
- Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S.; et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology 2022, 11, 275. [Google Scholar] [CrossRef]
- Sarrafi, A.; Gentzbittel, L. Genomics as Efficient Tools: Example Sunflower Breeding. Biotechnol. Agric. For. 2005, 55, 107–119. [Google Scholar] [CrossRef]
- Vear, F. Changes in Sunflower Breeding over the Last Fifty Years. OCL 2016, 23, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Meena, H.P.; Sujatha, M.; Varaprasad, K.S. Achievements and Bottlenecks of Heterosis Breeding of Sunflower (Helianthus Annuus L.) in India. Indian J. Genet. Plant Breed. 2013, 73, 123–130. [Google Scholar] [CrossRef]
- Britt, A.B.; Kuppu, S. Cenh3: An Emerging Player in Haploid Induction Technology. Front. Plant Sci. 2016, 7, 357. [Google Scholar] [CrossRef] [PubMed]
- Cvejić, S.; Radanović, A.; Dedić, B.; Jocković, M.; Jocić, S.; Miladinović, D. Genetic and Genomic Tools in Sunflower Breeding for Broomrape Resistance. Genes 2020, 11, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and Opportunities in Plant Breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef] [PubMed]
- Darqui, F.S.; Radonic, L.M.; Beracochea, V.C.; Hopp, H.E.; López Bilbao, M. Peculiarities of the Transformation of Asteraceae Family Species: The Cases of Sunflower and Lettuce. Front. Plant Sci. 2021, 12, 767459. [Google Scholar] [CrossRef] [PubMed]
- Dagustu, N. In Vitro Tissue Culture Studies in Sunflower (Helianthus Spp.). Ekin J. Crop Breed. Genet. 2018, 4, 13–21. [Google Scholar]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The Sunflower Genome Provides Insights into Oil Metabolism, Flowering and Asterid Evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [Green Version]
- Hübner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower Pan-Genome Analysis Shows That Hybridization Altered Gene Content and Disease Resistance. Nat. Plants 2019, 5, 54–62. [Google Scholar] [CrossRef]
- Ishii, T.; Karimi-Ashtiyani, R.; Houben, A. Haploidization via Chromosome Elimination: Means and Mechanisms. Annu. Rev. Plant Biol. 2016, 67, 421–438. [Google Scholar] [CrossRef]
- Murovec, J.; Bohanec, B.; Murovec, J.; Bohanec, B. Haploids and Doubled Haploids in Plant Breeding. Plant Breed. 2012, 2012, 87–106. [Google Scholar] [CrossRef] [Green Version]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point Mutation Impairs Centromeric CENH3 Loading and Induces Haploid Plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [Green Version]
- Eliby, S.; Bekkuzhina, S.; Kishchenko, O.; Iskakova, G.; Kylyshbayeva, G.; Jatayev, S.; Soole, K.; Langridge, P.; Borisjuk, N.; Shavrukov, Y. Developments and Prospects for Doubled Haploid Wheat. Biotechnol. Adv. 2022, 60, 108007. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Haploids in Flowering Plants: Origins and Exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wu, P.; Trampe, B.; Tian, X.; Lubberstedt, T.; Chen, S. Novel Technologies in Doubled Haploid Line Development. Plant Biotechnol. J. 2017, 15, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Jin, W.; Wang, K. Centromere Histone H3- and Phospholipase-Mediated Haploid Induction in Plants. Plant Methods 2019, 15, 42. [Google Scholar] [CrossRef] [Green Version]
- Drumeva, M.; Yankov, P. Parthenogenetic Responsiveness of Sunflower Hybrid Combinations with Expressed Tolerance to Herbicides. Agric. Sci. Technol. 2017, 9, 190–193. [Google Scholar] [CrossRef]
- Todorova, M.; Ivanov, P.; Shindrova, P.; Christov, M.; Ivanova, I. Doubled Haploid Production of Sunflower (Helianthus Annuus L.) through Irradiated Pollen-Induced Parthenogenesis. Euphytica 1997, 97, 249–254. [Google Scholar] [CrossRef]
- Bohorova, N. In Vitro Organogenesis, Androgenesis and Embryo Culture in the Genus Helianthus L. Z. Pflanzenzuchtg. 1985, 95, 35–44. [Google Scholar]
- Saji, K.V.; Sujatha, M. Embryogenesis and Plant Regeneration in Anther Culture of Sunflower (Helianthus Annuus L.). Euphytica 1998, 103, 1–7. [Google Scholar] [CrossRef]
- Jonard, R.; Mezzarobba, A. Sunflower (Helianthus Spp.): Anther Culture and Field Studies on Haploids; Springer: Berlin/Heidelberg, Germany, 1990; pp. 485–501. [Google Scholar] [CrossRef]
- Kucera, V.; Vyvadilov, M.; Klima, M. Utilisation of Doubled Haploids in Winter Oilseed Rape (Brassica Napus L.) Breeding. Czech J. Genet. Plant Breed. 2002, 38, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Ferrie, A.M.R.; Caswell, K.L. Applications of doubled haploidy for Improving industrial oilseeds. In Industrial Oil Crops; AOCS Press: Urbana, IL, USA, 2016; pp. 359–378. [Google Scholar] [CrossRef]
- Daurova, A.; Daurov, D.; Volkov, D.; Zhapar, K.; Raimbek, D.; Shamekova, M.; Zhambakin, K. Doubled Haploids of Interspecific Hybrids between Brassica Napus and Brassica Rapa for Canola Production with Valuable Breeding Traits. OCL 2020, 27, 45. [Google Scholar] [CrossRef]
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-Art and Novel Developments of in Vivo Haploid Technologies. Theor. Appl. Genet. 2019, 132, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Finch, R.A. Tissue-Specific Elimination of Alternative Whole Parental Genomes in One Barley Hybrid. Chromosoma 1983, 88, 386–393. [Google Scholar] [CrossRef]
- Piosik, Ł.; Zenkteler, E.; Zenkteler, M. Development of Haploid Embryos and Plants of Lactuca Sativa Induced by Distant Pollination with Helianthus Annuus and H. Tuberosus. Euphytica 2016, 208, 439–451. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a Sperm-Specific Phospholipase, Triggers Maize Haploid Induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Coumans, M.; Zhong, D. Doubled Haploid Sunflower (Helianthus Annuus) Plant Production by Androgenesis: Fact or Artifact? Part 2. In Vitro Isolated Microspore Culture. Plant Cell. Tissue Organ Cult. 1995, 41, 203–309. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-Step Genome Editing of Elite Crop Germplasm during Haploid Induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Sidhu, G.S.; Conner, J.A.; Ozias-Akins, P. Controlled Induction of Parthenogenesis in Transgenic Rice via Post-Translational Activation of PsASGR-BBML. Front. Plant Sci. 2022, 13, 925467. [Google Scholar] [CrossRef]
- Wang, N.; Gent, J.I.; Dawe, R.K. Haploid Induction by a Maize Cenh3 Null Mutant. Sci. Adv. 2021, 7, eabe2299. [Google Scholar] [CrossRef]
- Ravi, M.; Chan, S.W.L. Haploid Plants Produced by Centromere-Mediated Genome Elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere Identity from the DNA Point of View. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Nagaki, K.; Tanaka, K.; Yamaji, N.; Kobayashi, H.; Murata, M. Sunflower Centromeres Consist of a Centromere-Specific LINE and a Chromosome-Specific Tandem Repeat. Front. Plant Sci. 2015, 6, 912. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, S.; Ishii, T.; Brown, C.T.; Houben, A.; Comai, L. Centromere Location in Arabidopsis Is Unaltered by Extreme Divergence in CENH3 Protein Sequence. Genome Res. 2017, 27, 471–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamariola, L.; Tiang, C.L.; De Storme, N.; Pawlowski, W.; Geelen, D. Chromosome Segregation in Plant Meiosis. Front. Plant Sci. 2014, 5, 279. [Google Scholar] [CrossRef] [Green Version]
- Watts, A.; Singh, S.K.; Bhadouria, J.; Naresh, V.; Bishoyi, A.K.; Geetha, K.A.; Chamola, R.; Pattanayak, D.; Bhat, S.R. Brassica Juncea Lines with Substituted Chimeric GFP-CENH3 Give Haploid and Aneuploid Progenies on Crossing with Other Lines. Front. Plant Sci. 2017, 7, 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuppu, S.; Tan, E.H.; Nguyen, H.; Rodgers, A.; Comai, L. Point Mutations in Centromeric Histone Induce Post-Zygotic Incompatibility and Uniparental Inheritance. PLoS Genet. 2015, 11, e1005494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, M.; Marimuthu, M.P.A.; Tan, E.H.; Maheshwari, S.; Henry, I.M.; Marin-Rodriguez, B.; Urtecho, G.; Tan, J.; Thornhill, K.; Zhu, F.; et al. A Haploid Genetics Toolbox for Arabidopsis thaliana. Nat. Commun. 2014, 5, 5334. [Google Scholar] [CrossRef] [Green Version]
- Lermontova, I.; Koroleva, O.; Rutten, T.; Fuchs, J.; Schubert, V.; Moraes, I.; Koszegi, D.; Schubert, I. Knockdown of CENH3 in Arabidopsis Reduces Mitotic Divisions and Causes Sterility by Disturbed Meiotic Chromosome Segregation. Plant J. 2011, 68, 40–50. [Google Scholar] [CrossRef]
- Lv, J.; Yu, K.; Wei, J.; Gui, H.; Liu, C.; Liang, D.; Wang, Y.; Zhou, H.; Carlin, R.; Rich, R.; et al. Generation of Paternal Haploids in Wheat by Genome Editing of the Centromeric Histone CENH3. Nat. Biotechnol. 2020, 38, 1397–1401. [Google Scholar] [CrossRef]
- Wang, G.; Zong, M.; Liu, D.; Wu, Y.; Tian, S.; Han, S.; Guo, N.; Duan, M.; Miao, L.; Liu, F. Efficient Generation of Targeted Point Mutations in the Brassica Oleracea Var. Botrytis Genome via a Modified CRISPR/Cas9 System. Hortic. Plant J. 2022, 8, 527–530. [Google Scholar] [CrossRef]
- Kuppu, S.; Ron, M.; Marimuthu, M.P.A.; Li, G.; Huddleson, A.; Siddeek, M.H.; Terry, J.; Buchner, R.; Shabek, N.; Comai, L.; et al. A Variety of Changes, Including CRISPR/Cas9-Mediated Deletions, in CENH3 Lead to Haploid Induction on Outcrossing. Plant Biotechnol. J. 2020, 18, 2068–2080. [Google Scholar] [CrossRef]
- Muiruri, S.K. Development of Haploid Inducers in Musa spp. (Bananas) By Modification of Centromere Specific Histone 3 (CENH3) protein. Ph.D. Thesis, University of Nairobi, Nairobi, Kenya, 2015. [Google Scholar]
- Senthil-Kumar, M.; Mysore, K.S. RNAi in Plants: Recent Developments and Applications in Agriculture. Gene Silenc. Theory Tech. Appl. 2013, 183–200. [Google Scholar]
- Pradeep, K.; Satya, V.K.; Selvapriya, M.; Vijayasamundeeswari, A.; Ladhalakshmi, D.; Paranidharan, V.; Rabindran, R.; Samiyappan, R.; Balasubramanian, P.; Velazhahan, R. Engineering Resistance against Tobacco Streak Virus (TSV) in Sunflower and Tobacco Using RNA Interference. Biol. Plant. 2012, 56, 735–741. [Google Scholar] [CrossRef]
- Rajput, M.; Choudhary, K.; Kumar, M.; Vivekanand, V.; Chawade, A.; Ortiz, R.; Pareek, N. RNA Interference and CRISPR/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants 2021, 10, 1914. [Google Scholar] [CrossRef] [PubMed]
- Pacher, M.; Puchta, H. From Classical Mutagenesis to Nuclease-Based Breeding—Directing Natural DNA Repair for a Natural End-Product. Plant J. 2017, 90, 819–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathirana, R. Plant Mutation Breeding in Agriculture. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2011, 6, 1–20. [Google Scholar] [CrossRef]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M. Induced Mutagenesis in Plants Using Physical and Chemical Agents. Plant Cell Cult. Essent. Methods 2010, 20, 111–130. [Google Scholar] [CrossRef]
- Songstad, D.D.; Petolino, J.F.; Voytas, D.F.; Reichert, N.A. Genome Editing of Plants. Crit. Rev. Plant Sci. 2017, 36, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Fanelli, V.; Ngo, K.J.; Thompson, V.L.; Silva, B.R.; Tsai, H.; Sabetta, W.; Montemurro, C.; Comai, L.; Harmer, S.L. A TILLING by Sequencing Approach to Identify Induced Mutations in Sunflower Genes. Sci. Rep. 2021, 11, 9885. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [Green Version]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. The Problem of the Low Rates of CRISPR/Cas9-Mediated Knock-Ins in Plants: Approaches and Solutions. Int. J. Mol. Sci. 2019, 20, 3371. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, N.; Rahman, M.-U.; Mukhtar, Z.; Zafar, Y.; Zhang, B. A Critical Look on CRISPR-Based Genome Editing in Plants. J. Cell. Physiol. 2020, 235, 666–682. [Google Scholar] [CrossRef]
- Schiml, S.; Puchta, H. Revolutionizing Plant Biology: Multiple Ways of Genome Engineering by CRISPR/Cas. Plant Methods 2016, 12, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, C.; Yuan, J.; Wang, R.; Liu, Y.; Birchler, J.A.; Han, F. Efficient Targeted Genome Modification in Maize Using CRISPR/Cas9 System. J. Genet. Genom. 2016, 43, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas Immune System: Biology, Mechanisms and Applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The Emerging and Uncultivated Potential of CRISPR Technology in Plant Science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [Green Version]
- Schaeffer, S.M.; Nakata, P.A. The Expanding Footprint of CRISPR/Cas9 in the Plant Sciences. Plant Cell Rep. 2016, 35, 1451–1468. [Google Scholar] [CrossRef]
- Dunemann, F.; Krüger, A.; Maier, K.; Struckmeyer, S. Insights from CRISPR/Cas9-Mediated Gene Editing of Centromeric Histone H3 (CENH3) in Carrot (Daucus Carota Subsp. Sativus). bioRxiv 2022. [Google Scholar] [CrossRef]
- Yoon, S.; Bragg, J.; Aucar-Yamato, S.; Chanbusarakum, L.; Dluge, K.; Cheng, P.; Blumwald, E.; Gu, Y.; Tobias, C.M. Haploidy and Aneuploidy in Switchgrass Mediated by Misexpression of CENH3. Plant Genome 2022, e20209. [Google Scholar] [CrossRef] [PubMed]
- Stajič, E.; Kiełkowska, A.; Murovec, J.; Bohanec, B. Deep Sequencing Analysis of CRISPR/Cas9 Induced Mutations by Two Delivery Methods in Target Model Genes and the CENH3 Region of Red Cabbage (Brassica Oleracea Var. Capitata f. Rubra). Plant Cell Tissue Organ Cult. 2019, 139, 227–235. [Google Scholar] [CrossRef]
- Zaidi, S.S.-A.; Mansoor, S. Viral Vectors for Plant Genome Engineering. Front. Plant Sci. 2017, 8, 2012–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Wolt, J.D. Risk Associated with Off-Target Plant Genome Editing and Methods for Its Limitation. Emerg. Top. Life Sci. 2017, 1, 231–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, Y.; Liang, Z.; Gao, C. Current and Future Editing Reagent Delivery Systems for Plant Genome Editing. Sci. China Life Sci. 2017, 60, 490–505. [Google Scholar] [CrossRef]
- Altpeter, F.; Baisakh, N.; Beachy, R.; Bock, R.; Capell, T.; Christou, P.; Daniell, H.; Datta, K.; Datta, S.; Dix, P.J.; et al. Particle Bombardment and the Genetic Enhancement of Crops: Myths and Realities. Mol. Breed. 2005, 15, 305–327. [Google Scholar] [CrossRef]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.L.; Liu, Y. A Geminivirus-Based Guide RNA Delivery System for CRISPR/Cas9 Mediated Plant Genome Editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Wolter, F.; Puchta, H. Knocking out Consumer Concerns and Regulator’s Rules: Efficient Use of CRISPR/Cas Ribonucleoprotein Complexes for Genome Editing in Cereals. Genome Biol. 2017, 18, 43. [Google Scholar] [CrossRef] [Green Version]
- Zess, E.; Begemann, M. CRISPR-Cas9 and beyond: What’s next in Plant Genome Engineering. Vitr. Cell. Dev. Biol. Plant 2021, 57, 584–594. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-Faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef] [Green Version]
- Varanda, C.M.R.; Félix, M.D.R.; Campos, M.D.; Patanita, M.; Materatski, P. Plant Viruses: From Targets to Tools for Crispr. Viruses 2021, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Dinesh-Kumar, S.P.; Voytas, D.F. Editing through Infection. Nat. Plants 2020, 6, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Hooykaas, P.J.J.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Agrobacterium-Mediated Transformation as a Tool for Functional Genomics in Fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H.; Lomonossoff, G.P. When Plant Virology Met Agrobacterium: The Rise of the Deconstructed Clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellens, R.; Mullineaux, P.; Klee, H. A Guide to Agrobacterium Binary Ti Vectors. Trends Plant Sci. 2000, 5, 446–451. [Google Scholar] [CrossRef]
- Opabode, J.T. Agrobacterium-Mediated Transformation of Plants: Emerging Factors That Influence Efficiency. Biotechnol. Mol. Biol. Rev. 2006, 1, 12–20. [Google Scholar]
- Radonic, L.M.; Lewi, D.M.; López, N.E.; Hopp, H.E.; Escandón, A.S. Chapter 5 Sunflower (Helianthus Annuus L.). In Agrobacterium Protocols; Springer: New York, NY, USA, 2015; pp. 47–55. [Google Scholar] [CrossRef]
- Sujatha, M.; Vijay, S.; Vasavi, S.; Reddy, P.V.; Rao, S.C. Agrobacterium—Mediated Transformation of Cotyledons of Mature Seeds of Multiple Genotypes of Sunflower (Helianthus Annuus L.). Plant Cell Tissue Organ Cult. (PCTOC) 2012, 110, 275–287. [Google Scholar] [CrossRef]
- Heim, U.; Manteuffel, R.; Bäumlein, H.; Steinbiss, H.H.; Wobus, U. Transient Expression of a Lysine-Rich Vicilin Gene of Vicia Faba in Barley Endosperm Detected by Immunological Tissue Printing after Particle Bombardment. Plant Cell Rep. 1995, 15, 125–128. [Google Scholar] [CrossRef]
- Finer, J.J.; McMullen, M.D. Transformation of Soybean via Particle Bombardment of Embryogenic Suspension Culture Tissue. Vitr. Cell. Dev. Biol.Plant 1991, 27, 175–182. [Google Scholar] [CrossRef]
- Hunold, R.; Bronner, R.; Hahne, G. Early Events in Microprojectile Bombardment: Cell Viability and Particle Location. Plant J. 1994, 5, 593–604. [Google Scholar] [CrossRef]
- Lucas, O.; Kallerhoff, J.; Alibert, G. Production of Stable Transgenic Sunflowers (Helianthus Annuus L.) from Wounded Immature Embryos by Particle Bombardment and Co-Cultivation with Agrobacterium tumefaciens. Mol. Breed. 2000, 6, 479–487. [Google Scholar] [CrossRef]
- Knittel, N.; Gruber, V.; Hahne, G.; Lénée, P. Transformation of Sunflower (Helianthus Annuus L.): A Reliable Protocol. Plant Cell Rep. 1994, 14, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Peretz, Y.; Mozes-koch, R.; Akad, F.; Tanne, E.; Czosnek, H.; Sela, I. A Universal Expression/Silencing Vector in Plants. Plant Physiol. 2007, 145, 1251–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivprasad, S.; Pogue, G.P.; Lewandowski, D.J.; Hidalgo, J.; Donson, J.; Grill, L.K.; Dawson, W.O. Heterologous Sequences Greatly Affect Foreign Gene Expression in Tobacco Mosaic Virus-Based Vectors. Virology 1999, 255, 312–323. [Google Scholar] [CrossRef] [Green Version]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato Virus X as a Vector for Gene Expression in Plants. Plant J. 1992, 2, 549–557. [Google Scholar] [CrossRef]
- Oh, Y.; Kim, H.; Kim, S. Virus-Induced Plant Genome Editing. Curr. Opin. Plant Biol. 2021, 60, 101992. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Viral Vectors for the Expression of Proteins in Plants. Curr. Opin. Biotechnol. 2007, 18, 134–141. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved Vectors and Genome-Wide Libraries for CRISPR Screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [Green Version]
- Eini, O.; Schumann, N.; Niessen, M.; Varrelmann, M. Targeted Mutagenesis in Plants Using Beet Curly Top Virus for Efficient Delivery of CRISPR/Cas12a Components. N. Biotechnol. 2022, 67, 1–11. [Google Scholar] [CrossRef]
- Rezaei, A.; Farsi, M.; Malekzadeh-Shafaroudi, S.; Seifi, A. In Planta Removal of NptII Selectable Marker Gene from Transgenic Tobacco Plants Using CRISPR/Cas9 System. Plant Gene 2021, 26, 100288. [Google Scholar] [CrossRef]
- Hayes, R.J.; Coutts, R.H.A.; Buck, K.W. Stability and Expression of Bacterial Genes in Replicating Geminivirus Vectors in Plants. Nucleic Acids Res. 1989, 17, 2391–2403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drumeva, M.; Yankov, P.; Nenova, N.; Shindrova, P.; Encheva, V. New Sunflower Restorer Lines Developed by Y-Induced Parthenogenesis from Helianthus Annuus Hybrids—Disease Resistance, Combining Ability. I. Disease Resistance. In Proceedings of the Conventional and Molecular Breeding of Field and Vegetable Crops, Novi Sad, Serbia, 24–27 November 2008. [Google Scholar] [CrossRef]
- Jin, K.; Tian, N.; da Silva Ferreira, J.F.; Sandhu, D.; Xiao, L.; Gu, M.; Luo, Y.; Zhang, X.; Liu, G.; Liu, Z.; et al. Comparative Transcriptome Analysis of Agrobacterium tumefaciens Reveals the Molecular Basis for the Recalcitrant Genetic Transformation of Camellia sinensis L. Biomolecules 2022, 12, 688. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G. The Way to True Plant Genome Editing. Nat. Plants 2020, 6, 736–737. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Daros, J. Tools and Targets: The Dual Role of Plant Viruses in CRISPR–Cas. Plant Genom 2022, e20220. [Google Scholar] [CrossRef]

| Plant Species | Haploid Induction Method | Application | References |
|---|---|---|---|
| Sunflower (H. annuus) | Parthenogenesis | Resistance to broomrape, phoma, imidazolinone, and downy mildew | Drumeva [5,25]; Todorova [26] |
| Anther culture | Fertility restoration | Bohorova [27]; Saji and Sujatha [28]; Jonard and Mezzarobba [29] | |
| Brassica napus L. (Rapeseed) | Spontaneously occurring | Oil yield enhancement | Reviewed by Kucera et al. [30] |
| Brassica spp. | Microspore culture/embryogenesis | Modification of fatty acid profiles, fatty acid levels, and drought tolerance | Reviewed by Ferrie et al. [31];Daurova et al. [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mabuza, L.M.; Mchunu, N.P.; Crampton, B.G.; Swanevelder, D.Z.H. Accelerated Breeding for Helianthus annuus (Sunflower) through Doubled Haploidy: An Insight on Past and Future Prospects in the Era of Genome Editing. Plants 2023, 12, 485. https://doi.org/10.3390/plants12030485
Mabuza LM, Mchunu NP, Crampton BG, Swanevelder DZH. Accelerated Breeding for Helianthus annuus (Sunflower) through Doubled Haploidy: An Insight on Past and Future Prospects in the Era of Genome Editing. Plants. 2023; 12(3):485. https://doi.org/10.3390/plants12030485
Chicago/Turabian StyleMabuza, Londiwe M., Nokuthula P. Mchunu, Bridget G. Crampton, and Dirk Z. H. Swanevelder. 2023. "Accelerated Breeding for Helianthus annuus (Sunflower) through Doubled Haploidy: An Insight on Past and Future Prospects in the Era of Genome Editing" Plants 12, no. 3: 485. https://doi.org/10.3390/plants12030485




