Investigation of Cannabis sativa Phytochemicals as Anti-Alzheimer’s Agents: An In Silico Study
Abstract
:1. Introduction
2. Results
2.1. Drug Likeliness Properties Analysis
2.2. ADMET Analysis
2.3. Toxicity Analysis
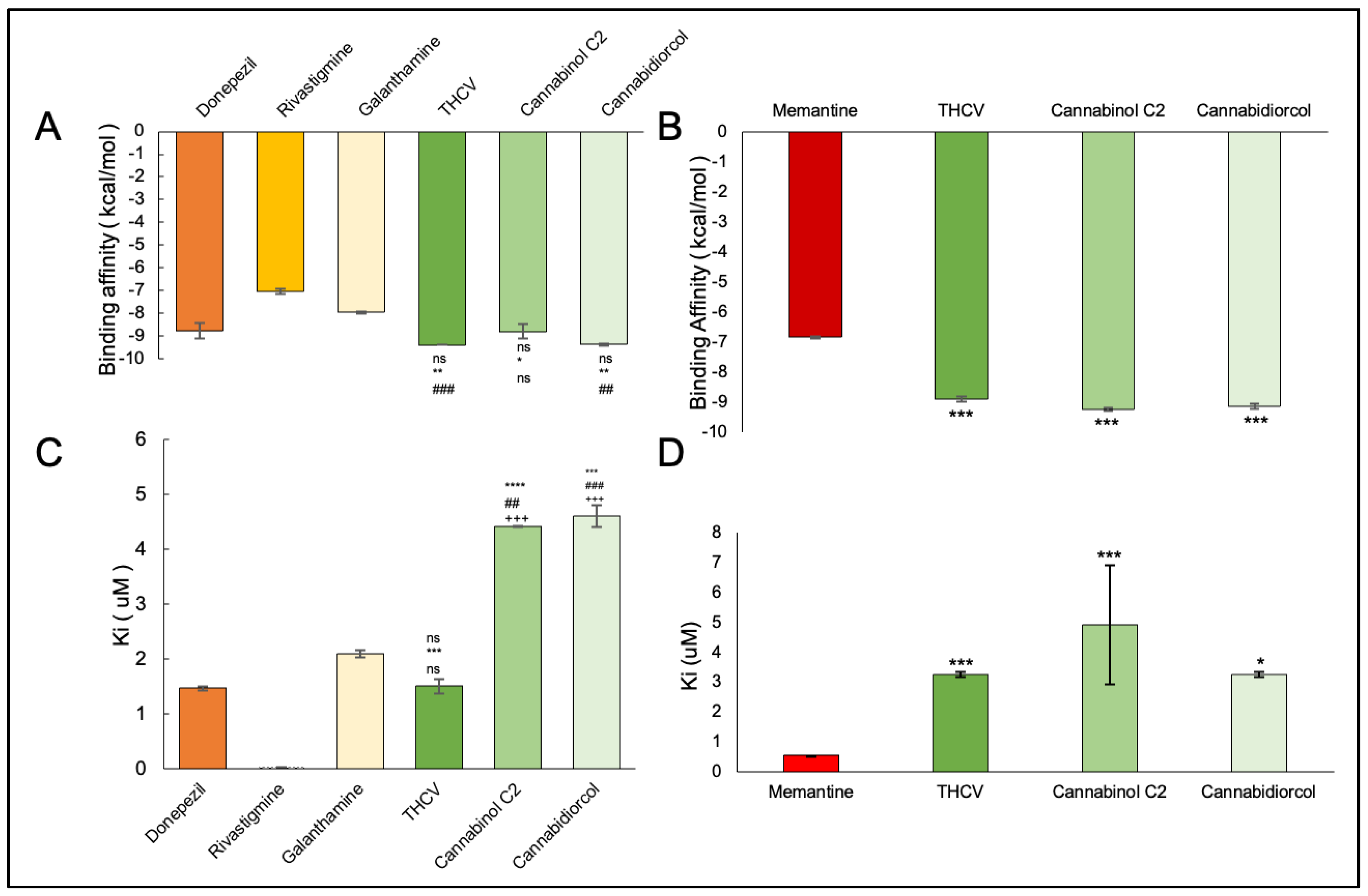
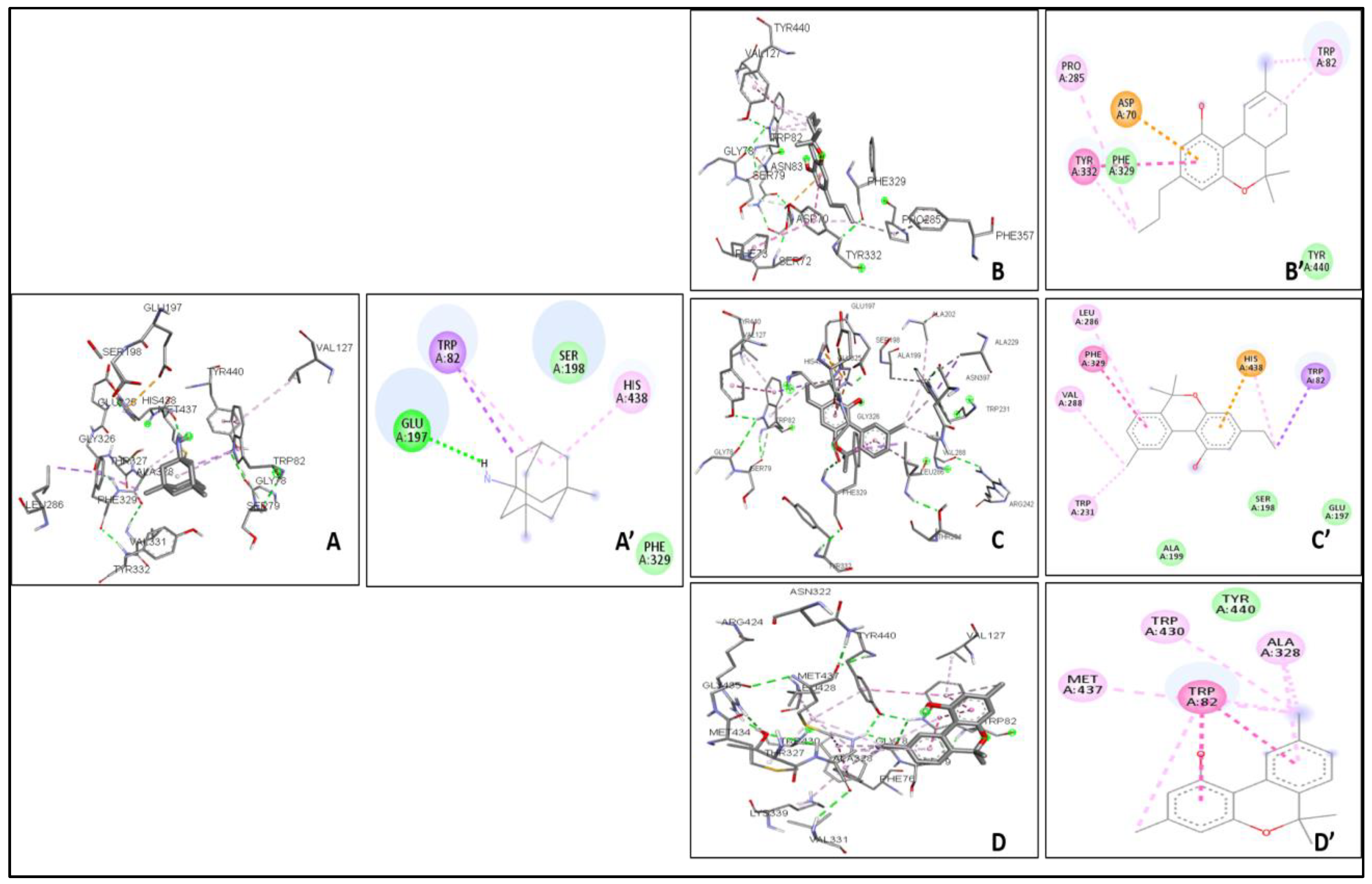
2.4. Molecular Docking
3. Discussion
4. Materials & Methods
4.1. Retrieval of the Ligand Molecule and Protein Structure for ADME Studies
4.1.1. Protein Preparation
4.1.2. Ligand Preparation
4.2. Drug Likeliness Properties
4.3. ADME and Toxicity Test
4.3.1. ADME Properties
4.3.2. Toxicity Prediction
4.4. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CBD | Cannabidiol |
| THC | Tetrahydrocannabinol |
| AChE | Acetylcholinesterase |
| BuChE | Butryrylcholinesterase |
| AD | Alzheimer’s disease |
| γ-secretase | Gamma-secretase |
| BACE-1 | Beta secretase-1 |
| ADMET | Absorption, Distribution, Metabolism, Excretion and Toxicity |
| THCV | Tetrahydrocannabivarin |
| iGEMDOCK | A Graphical Environment for Recognizing Pharmacological Interactions and Virtual Screening |
| GPR55 | G protein-coupled receptors 55 |
| GPR3 | G protein-coupled receptors 3 |
| NF-κB | nuclear factor κB |
| PPARγ | peroxisome proliferator-activated receptor γ |
| GPCRs | G protein-coupled receptors |
| BBB | Blood brain barrier |
| APP | Amyloid precursor protein |
| GPP | geranyl diphosphate |
| OLA | olivetolic acid |
| NFT | neurofibilliary tangles |
| Aβ | beta-amyloid |
| sAPP-β | soluble amyloid precursor protein β |
| CYP2D6 | Cytochrome P450 2D6 |
| CYP3A4 | Cytochrome P450 3A4 |
| P450s | cytochrome P450 enzymes |
| hERG | human ether-a-go-go-related gene |
| LD50 | lethal dose 50 |
| VDW | van der Waals interactions |
| HB | hydrogen bonds |
| SHY5YAPP+ cells | SH-SY5Y cells transfected with the amyloid precursor protein |
| CASTp | Computed Atlas of Surface Topography of proteins |
| Ki | inhibition constant of enzymes |
| ΔG | Gibbs free energy |
| TE | electrostatic forces |
| Kcal/mol | kilo-calorie per mole |
| PAINS | Pan Assay Interference Compounds |
| TPSA | topological molecular polar surface area |
| ClogP | log(coctanol/cwater) |
| HIA | human gastrointestinal absorption |
References
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Dowling, C.A.; Melzer, R.; Schilling, S. Timing is everything: The genetics of flowering time in Cannabis sativa. Biochemist 2021, 43, 34–38. [Google Scholar] [CrossRef]
- Andre, C.M.; Hausman, J.F.; Guerriero, G. Cannabis sativa: The plant of the thousand and one molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banister, S.D.; Arnold, J.C.; Connor, M.; Glass, M.; McGregor, I.S. Dark classics in chemical neuroscience: Δ9-tetrahydrocannabinol. ACS Chem. Neurosci. 2019, 10, 2160–2175. [Google Scholar] [CrossRef]
- Gill, E.W.; Paton, W.D.M.; Pertwee, R.G. Preliminary Experiments on the Chemistry and Pharmacology of Cannabis. Nature 1970, 228, 134–136. [Google Scholar] [CrossRef]
- McPartland, J.M.; Russo, E.B. Cannabis and Cannabis Extracts: Greater Than the Sum of Their Parts? J. Cannabis Ther. 2001, 1, 103–132. [Google Scholar] [CrossRef]
- Chandra, S.; Lata, H.; ElSohly, M.A. Cannabis sativa L.-Botany and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 3-319-54564-7. [Google Scholar]
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A Unified Critical Inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [Green Version]
- Citti, C.; Linciano, P.; Panseri, S.; Vezzalini, F.; Forni, F.; Vandelli, M.A.; Cannazza, G. Cannabinoid Profiling of Hemp Seed Oil byLiquid Chromatography Coupled to High-Resolution Mass Spectrometry. Front. Plant Sci. 2019, 10, 120. [Google Scholar] [CrossRef]
- Pavlovic, R.; Panseri, S.; Giupponi, L.; Leoni, V.; Citti, C.; Cattaneo, C.; Cavaletto, M.; Giorgi, A. Phytochemical and EcologicalAnalysis of Two Varieties of Hemp (Cannabis sativa L.) Grown in a Mountain Environment of Italian Alps. Front. Plant Sci. 2019, 10, 1265. [Google Scholar] [CrossRef]
- Pertwee, R.G. Cannabinoid pharmacology: The first 66 years. Br. J. Pharmacol. 2006, 147, 163–171. [Google Scholar] [CrossRef]
- Curran, H.V.; Freeman, T.P.; Mokrysz, C.; Lewis, D.A.; Morgan, C.J.A.; Parsons, L.H. Keep off the grass? Cannabis, cognition and addiction. Nat. Rev. Neurosci. 2016, 17, 293–306. [Google Scholar] [CrossRef] [PubMed]
- ElSohly, M.A.; Radwan, M.M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of Cannabis sativa L. In Progress in the Chemistry of Organic Natural Products; Kinghorn, A., Falk, H., Gibbons, S., Kobayashi, J., Eds.; Springer: Cham, Switzerland, 2017; Volume 103, pp. 1–36. [Google Scholar]
- Lewis, M.M.; Yang, Y.; Wasilewski, E.; Clarke, H.A.; Kotra, L.P. Chemical profiling of medical cannabis extracts. ACS Omega 2017, 2, 6091–6103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Borah, R.; Sharma, B.; Pandhi, S.; Tripathi, V.; Yadav, H.S.; Devi, S.; Patil, U.; et al. Pharmacological properties, therapeutic potential, and legal status of Cannabis sativa L.: An overview. Phytother. Res. 2021, 35, 6010–6029. [Google Scholar] [CrossRef]
- Abuhasira, R.; Schleider, L.B.L.; Mechoulam, R.; Novack, V. Epidemiological characteristics, safety and efficacy of medical cannabis in the elderly. Eur. J. Intern. Med. 2018, 49, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.C.; Tsien, R.W.; Whalley, B.J.; Devinsky, O. Cannabinoids and epilepsy. Neurotherapeutics 2015, 12, 747–768. [Google Scholar] [CrossRef] [Green Version]
- Nuutinen, T. European journal of medicinal chemistry medicinal properties of terpenes found in cannabis sativa and Humulus lupulus. Eur. J. Med. Chem. 2018, 157, 198–228. [Google Scholar] [CrossRef] [PubMed]
- Aizpurua-Olaizola, O.; Soydaner, U.; Oztürk, Ë.; Schibano, D.; Simsir, Y.; Navarro, P.; Etxebarria, N.; Usobiaga, A. Evolution of the cannabinoid and terpene content during the growth of cannabis sativa plants from different chemotypes. J. Nat. Prod. 2016, 79, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Shekher, A.; Puneet Narula, A.S.; Abrahamse, H.; Gupta, S.C. Cannabis and its constituents for cancer: History, biogenesis, chemistry and pharmacological activities. Pharmacol. Res. 2021, 163, 105302. [Google Scholar] [CrossRef] [PubMed]
- Iuvone, T.; Esposito, G.; Esposito, R.; Santamaria, R.; Di Rosa, M.; Izzo, A.A. Neuroprotective effect of cannabidiol, a non-psychoactive component from Cannabis sativa, on β-amyloid-induced toxicity in PC12 cells. J. Neurochem. 2004, 89, 134–141. [Google Scholar] [CrossRef]
- Esposito, G.; De Filippis, D.; Carnuccio, R.; Izzo, A.A.; Iuvone, T. The marijuana component cannabidiol inhibits beta-amyloid-induced tau protein hyperphosphorylation through Wnt/beta-catenin pathway rescue in PC12 cells. J. Mol. Med. 2006, 84, 253–258. [Google Scholar] [CrossRef]
- Eubanks, L.M.; Rogers, C.J.; Beuscher, A.; Koob, G.F.; Olson, A.J.; Dickerson, T.J.; Janda, K.D. A molecular link between the active component of marijuana and Alzheimer’s disease pathology. Mol. Pharm. 2006, 3, 773–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassano, T.; Villani, R.; Pace, L.; Carbone, A.; Bukke, V.N.; Orkisz, S.; Avolio, C.; Serviddio, G. From Cannabis sativa to cannabidiol: Promising therapeutic candidate for the treatment of neurodegenerative diseases. Front. Pharmacol. 2020, 6, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuñez-Borque, E.; González-Naranjo, P.; Bartolomé, F.; Alquézar, C.; Reinares-Sebastián, A.; Pérez, C.; Ceballos, M.L.; Páez, J.A.; Campillo, N.E.; Martín-Requero, Á. Targeting cannabinoid receptor activation and BACE-1 activity counteracts TgAPP mice memory impairment and Alzheimer’s disease lymphoblast alterations. Mol. Neurobiol. 2020, 57, 1938–1951. [Google Scholar] [CrossRef] [PubMed]
- Citti, C.; Linciano, P.; Russo, F.; Luongo, L.; Iannotta, M.; Maione, S.; Laganà, A.; Capriotti, A.L.; Forni, F.; Vandelli, M.A.; et al. A novel phytocannabinoid isolated from Cannabis sativa L. with an in vivo cannabimimetic activity higher than Δ9-tetrahydrocannabinol: Δ9-Tetrahydrocannabiphorol. Sci. Rep. 2019, 9, 20335. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zhang, J.; Fan, N.; Teng, Z.; Wu, Y.; Yang, H.; Tang, Y.; Sun, H.; Song, Y.; Chen, C. ∆9-THC-Caused Synaptic and Memory Impairments Are Mediated through COX-2 Signaling. Cell 2013, 155, 1154–1165. [Google Scholar] [CrossRef] [Green Version]
- Franke, T.N.; Irwin, C.; Beindorff, N.; Bouter, Y.; Bouter, C. Effects of tetrahydrocannabinol treatment on brain metabolism and neuron loss in a mouse model of sporadic Alzheimer’s disease. Nuklearmedizin-Nucl. 2019, 58, P94. [Google Scholar]
- Wiles, D.; Shanbhag, B.K.; O’Brien, M.; Doblin, M.S.; Bacic, A.; Beddoe, T. Heterologous production of Cannabis sativa-derived specialised metabolites of medicinal significance–Insights into engineering strategies. Phytochemistry 2022, 203, 113380. [Google Scholar] [CrossRef]
- Schubert, D.; Kepchia, D.; Liang, Z.; Dargusch, R.; Goldberg, J.; Maher, P. Efficacy of cannabinoids in a pre-clinical drug-screening platform for Alzheimer’s disease. Mol. Neurobiol. 2019, 56, 7719–7730. [Google Scholar] [CrossRef]
- Shinjyo, N.; Di Marzo, V. The effect of cannabichromene on adult neural stem/progenitor cells. Neurochem. Int. 2013, 63, 432–437. [Google Scholar] [CrossRef]
- Werz, O.; Seegers, J.; Schaible, A.M.; Weinigel, C.; Barz, D.; Koeberle, A.; Appendino, G. Cannflavins from hemp sprouts, a novel cannabinoid-free hemp food product, target microsomal prostaglandin E2 synthase-1 and 5-lipoxygenase. PharmaNutrition 2014, 2, 53–60. [Google Scholar] [CrossRef]
- Colović, M.B.; Krstić, D.Z.; Lazarević-Pašti, T.D.; Bondžić, A.M.; Vasić, V.M. Acetylcholinesterase inhibitors: Pharmacology and toxicology. Curr. Neuropharmacol. 2013, 11, 315–335. [Google Scholar] [CrossRef] [PubMed]
- Djurfeldt, M.; Hjorth, J.; Eppler, J.M.; Dudani, N.; Helias, M.; Potjans, T.C.; Ekeberg, Ö. Run-time interoperability between neuronal network simulators based on the MUSIC framework. Neuroinformatics 2010, 8, 43–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espadas, I.; Keifman, E.; Palomo-Garo, C.; Burgaz, S.; García, C.; Fern’ andez-Ruiz, J.; Moratalla, R. Beneficial effects of the phytocannabinoid Δ9-THCV in L-DOPAinduced dyskinesia in Parkinson’s disease. Neurobiol. Dis. 2020, 141, 104892. [Google Scholar] [CrossRef]
- Rea, K.A.; Casaretto, J.A.; Al-Abdul-Wahid, M.S.; Sukumaran, A.; Geddes-McAlister, J.; Rothstein, S.J.; Akhtar, T.A. Biosynthesis of cannflavins A and B from Cannabis sativa L. Phytochemistry 2019, 164, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.L.; Gordon, D.; Evans, F.J. Isolation from Cannabis sativa L. of cannflavin—A novel inhibitor of prostaglandin production. Biochem. Pharmacol. 1985, 34, 2019–2024. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, S.; Kolishetti, N.; Arias, A.Y.; Vashist, A.; Nair, M. Cannabidiol for neurodegenerative disorders: A comprehensive review. Front. Pharmacol. 2022, 13, 989717. [Google Scholar] [CrossRef] [PubMed]
- Coles, M.; Steiner-Lim, G.Z.; Karl, T. Therapeutic properties of multi-cannabinoid treatment strategies for Alzheimer’s disease. Front. Neurosci. 2022, 16, 962922. [Google Scholar] [CrossRef]
- Tiwari, S.; Atluri, V.; Kaushik, A.; Yndart, A.; Nair, M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. Int. J. Nanomed. 2019, 14, 5541. [Google Scholar] [CrossRef] [Green Version]
- Alzheimer’s Association. 2021 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2021, 17, 327–406. [Google Scholar] [CrossRef]
- Bortolato, B.; Miskowiak, K.W.; Köhler, C.A.; Maes, M.; Fernandes, B.S.; Berk, M.; Carvalho, A.F. Cognitive remission: A novel objective for the treatment of major depression? BMC Med. 2016, 14, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Menting, K.W.; Claassen, J.A. β-secretase inhibitor; a promising novel therapeutic drug in Alzheimer’s disease. Front. Aging Neurosci. 2014, 6, 165. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Tang, J.; dos Santos Passos, C.; Nurisso, A.; SimõesPires, C.A.; Ji, M.; Lou, H.; Fan, P. Characterization of Lignanamides from hemp (Cannabis sativa L.) seed and their antioxidant and acetylcholinesterase inhibitory activities. J. Agric. Food Chem. 2015, 63, 10611–10619. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.S.; Jhanji, N. Natural products as potential anti-Alzheimer agents. Curr. Med. Chem. 2020, 27, 5887–5917. [Google Scholar] [CrossRef]
- Hanseeuw, B.J.; Betensky, R.A.; Jacobs, H.I.L.; Schultz, A.P.; Sepulcre, J.; Becker, J.A.; Cosio, D.M.O.; Farrell, M.; Quiroz, Y.T.; Mormino, E.C.; et al. Association of amyloid and tau with cognition in preclinical Alzheimer disease: A longitudinal study. JAMA Neurol. 2019, 76, 915–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.; Castro, A. Novel cholinesterase inhibitors as future effective drugs for the treatment of Alzheimer’s disease. Expert Opin. Investig. Drugs 2006, 15, 1–12. [Google Scholar] [CrossRef]
- Alzheimer’s Association Report. Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 11, 1–88.
- Masondo, N.A.; Stafford, G.I.; Aremu, A.O.; Makunga, N.P. Acetylcholinesterase inhibitors from southern African plants: An overview of ethnobotanical, pharmacological potential and phytochemical research including and beyond Alzheimer’s disease treatment. S. Afr. J. Bot. 2019, 120, 39–64. [Google Scholar] [CrossRef]
- Paunescu, H.; Dima, L.; Ghita, I.; Coman, L.; Ifteni, P.I.; Fulga, I.; Coman, O.A. A systematic review of clinical studies on the effect of psychoactive cannabinoids in psychiatric conditions in Alzheimer Dementia. Am. J. Ther. 2020, 27, e249–e269. [Google Scholar] [CrossRef]
- Holst, B.; Williamson, G. Nutrients and phytochemicals: From bioavailability to bioefficacy beyond antioxidants. Curr. Opin. Biotech. 2008, 19, 73–82. [Google Scholar] [CrossRef]
- Campbell, V.A.; Gowran, A. Alzheimer’s disease; taking the edge off with cannabinoids? Brit. J. Pharmacol. 2007, 152, 655–662. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Lim, C.S. Understanding the Modulatory Effects of Cannabidiol on Alzheimer’s Disease. Brain Sci. 2021, 11, 1211. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Feng, Y. Cannabidiol (CBD) enhanced the hippocampal immune response and autophagy of APP/PS1 Alzheimer’s mice uncovered by RNA-seq. Life Sci. 2021, 264, 118624. [Google Scholar] [CrossRef] [PubMed]
- Sirbu, C.A.; Manole, A.M.; Vasile, T.M.; Toma, G.S.; Dobrican, L.R.; Vîrvara, D.G.; Vasiliu, O. Cannabinoids—A new therapeutic strategy in neurology. Rom. J. Mil. Med. 2022, 125, 349. [Google Scholar] [CrossRef]
- Ibeas Bih, C.; Chen, T.; Nunn, A.V.; Bazelot, M.; Dallas, M.; Whalley, B.J. Molecular targets of cannabidiol in neurological disorders. Neurotherapeutics 2015, 12, 699–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Zhang, Y.; Chi, P.; Liu, H.; Jing, Z.; Cao, H.; Du, Y.; Zhao, Y.; Qin, X.; Zhang, W.; et al. Essential Oils: Chemical Constituents, Potential Neuropharmacological Effects and Aromatherapy-A Review. Pharmacol. Res. Mod. Chin. Med. 2022, 6, 100210. [Google Scholar] [CrossRef]
- Komorowska-Müller, J.A.; Schmöle, A.C. CB2 receptor in microglia: The guardian of self-control. Int. J. Mol. Sci. 2020, 22, 19. [Google Scholar] [CrossRef]
- Vallée, A.; Lecarpentier, Y.; Guillevin, R.; Vallée, J.N. Effects of cannabidiol interactions with Wnt/β-catenin pathway and PPARγ on oxidative stress and neuroinflammation in Alzheimer’s disease. Acta Biochim. Biophys. Sin. 2017, 49, 853–866. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Yang, J.W.; Kim, K.H.; Kim, J.U.; Yook, T.H. A Review on Studies of Marijuana for Alzheimer’s Disease–Focusing on CBD, THC. J. Pharmacopunct. 2019, 22, 225. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Z.; Zuo, J.; Wu, C.; Zha, L.; Xu, Y.; Wang, S.; Shi, J.; Liu, X.H.; Zhang, J.; et al. Novel cannabidiol—Carbamate hybrids as selective BuChE inhibitors: Docking-based fragment reassembly for the development of potential therapeutic agents against Alzheimer’s disease. Eur. J. Med. Chem. 2021, 223, 113735. [Google Scholar] [CrossRef]
- Makhouri, F.R.; Ghasemi, J.B. In silico studies in drug research against neurodegenerative diseases. Curr. Neuropharmacol. 2018, 16, 664–725. [Google Scholar] [CrossRef]
- Hill, A.J.; Weston, S.E.; Jones, N.A.; Smith, I.; Bevan, S.A.; Williamson, E.M.; Stephens, G.J.; Williams, C.M.; Whalley, B.J. Δ9-Tetrahydrocannabivarin suppresses in vitro epileptiform and in vivo seizure activity in adult rats. Epilepsia 2010, 51, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- García, C.; Palomo-Garo, C.; García-Arencibia, M.; Ramos, J.A.; Pertwee, R.G.; Fernández-Ruiz, J. Symptom-relieving and neuroprotective effects of the phytocannabinoid Δ9-THCV in animal models of Parkinson’s disease. Br. J. Pharmacol. 2011, 163, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Baroi, S.; Saha, A.; Bachar, R.; Bachar, S.C. Cannabinoid as potential aromatase inhibitor through molecular modeling and screening for anti-cancer activity. Dhaka Univ. J. Pharm. Sci. 2020, 26, 47–58. [Google Scholar] [CrossRef]
- Zoltán, O.; László, P.; Éva, K.; Béla, V. A “keto-enol” plaque buster mechanism to diminish Alzheimer’s β-Amyloid burden. Biochem. Biophys. Res. Comm. 2020, 532, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Scuderi, C.; Steardo, L.; Esposito, G. Cannabidiol promotes amyloid precursor protein ubiquitination and reduction of beta amyloid expression in SHSY5YAPP+ cells through PPARγ involvement. Phytother. Res. 2014, 28, 1007–1013. [Google Scholar] [CrossRef]
- Ramachandran, S.; Kota, P.; Ding, F.; Dokholyan, N.V. Homology modeling: Generating structural models to understand protein function and mechanism. InComputational modeling of biological systems. PROTEINS Struct. Funct. Bioinform. 2011, 79, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Patel, N.B.; Patel, L.N.; Patel, K.D.; Patel, M.V.; Kalasariya, H.S. Admet & Cytotoxicity Prediction of Red Seaweed Gracillaria Dura: An in Silico Approach. Available online: https://www.researchgate.net/profile/Nikunj-Patel-22/publication/339973699_ADMET_CYTOTOXICITY_PREDICTION_OF_RED_SEAWEED_GRACILLARIA_DURA_AN_IN_SILICO_APPROACH/links/5e708cdd92851c1a689a7c90/ADMET-CYTOTOXICITY-PREDICTION-OF-RED-SEAWEED-GRACILLARIA-DURA-AN-IN-SILICO-APPROACH.pdf (accessed on 29 November 2022).
- Hussain, H.; Ahmad, S.; Shah, S.W.; Ghias, M.; Ullah, A.; Rahman, S.U.; Kamal, Z.; Khan, F.A.; Khan, N.M.; Muhammad, J.; et al. Neuroprotective Potential of Synthetic Mono-Carbonyl Curcumin Analogs Assessed by Molecular Docking Studies. Molecules 2021, 26, 7168. [Google Scholar] [CrossRef]
- Van De Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucl. Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.C.; Gomes, T.M.; Pinto, B.A.; Camara, A.L.; Paes, A.M. Naturally occurring acetylcholinesterase inhibitors and their potential use for Alzheimer’s disease therapy. Front. Pharmacol. 2018, 18, 1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folch, J.; Busquets, O.; Ettcheto, M.; Sánchez-López, E.; Castro-Torres, R.D.; Verdaguer, E.; Garcia, M.L.; Olloquequi, J.; Casadesús, G.; Beas-Zarate, C.; et al. Memantine for the treatment of dementia: A review on its current and future applications. J. Alz. Dis. 2018, 62, 1223–1240. [Google Scholar] [CrossRef] [PubMed]



| Compound Class & Plant Tissue Type | Name | Effects on AD (In Vivo) | Effects on AD (In Vitro) | Precursor | Medicinal Characteristics [3,18] | References |
|---|---|---|---|---|---|---|
| Neutral cannabinoids (Trichomes, Female flowers, Roots/Apoplast (secretion pathway)) | Cannabidiol (CBD) | Male Wistar rats utilise it as a Streptozotocin (STZ)- induced AD model, CBD enhances the brain glucose metabolism. Activation of the PPARγ via Wnt/β-catenin pathway | Pretreatment restores the synaptic transmission that was reduced by Aβ in a C57 mouse hippocampal slice. | CBGA | Anti–fungal and anti–bacterial against methicillin resistant strains, sedative and analgesic potential and anti–epileptic potential | [19,20,21,22,23,24,25] |
| Tetrahydrocannabinol(THC) | THC reduces the Aβ burden in 5XFAD/APP mice | Compared to untreated controls, transgenic Tg4-42 mice expressing human A4-42 showed less neuronal death. | CBGA | Psychotropic & psychoactive properties | [20,26,27,28] | |
| Cannabichromene (CBC) | CBC (10–75 mgkg−1 i.p. per day) significantly decreased motor activity in a model of electroshock seizure during the first 10 min interval, but only the maximum dose was beneficial. | in vitro CBC improved the viability of neural stem cells | CBGA | Anti–inflammatory, sedative and analgesic potential | [29,30,31,32] | |
| Cannabidivarin (CBDV) | Inhibits oxytosis and prevents loss of energy in HT22 cells (50% inhibition at 1.1 μM and 90 nM, respectively), as well as reducing Aβ toxicity (50% inhibition at 100 nM) and trophic withdrawal (50% inhibition 350 nM); | Prevents oxytosis in Ht22 cells (mouse hippocampal cell) MC65 cells (human nerve cell line) | CBGVA | _ | [30,33,34] | |
| Cannabicyclol (CBL) | _ | _ | CBC | _ | [20,33] | |
| Cannabinol (CBN) | Inhibiting oxytosis and prevent loss of energy in HT22 cells (50% inhibition at 1.1 μM and 90 nM, respectively), as well as reducing Aβ toxicity (50% inhibition at 100 nM) and trophic withdrawal (50% inhibition 350 nM); | Along with its capacity to promote the breakdown and clearance of pre-formed A aggregates in MC65 cells at a concentration of 100 nM in HT-22 cells and cortical embryonic E18 neurons | THC | Mild psychoactive potential | [30,33] | |
| Cannabidiphorol (CBDP) | _ | _ | _ | Antinociceptive, | [35] | |
| Tetrahydrocannabivarin (THCV) | _ | _ | CBGVA | Anti–dyskinesia in Parkinson’s disease | [35] | |
| Flavonols (leaves, stems, seeds/lypophyl nature suggest cellular retention) | Cannabigerol (CBG) | Retains trophic factors present in cortical neurons of rat (effective concentration 50% = 1.5 μM) and inhibits the oxytosis in nerve cells (HT22) of mouse | Prevents oxytosis in Ht22 cells (mouse hippocampal cell MC65 cells (human nerve cell line) | OLA, GPP | Analgesic, Anti–inflammatory, Anti–Cancer, Psychotropic, Psychoactive | [30,34,36] |
| Canniflavin A | _ | Exhibits anti-inflammatory activity | Chrysoeriol | _ | [32,36,37] |
| Compound | Mw | HA | HD | Absorption | Lipinski’s Rule Violation | Solubility | BBB Permeability | CNS Permeability | CYP2D6 | LD50 (mg/kg) | Toxicity Class |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cannabidiorcol | 254.3 | 2 | 1 | 92.851 | 0 | −4.599 | 0.397 | −1.368 | NO | 800 | 4 |
| THCV | 286.4 | 2 | 1 | 91.821 | 0 | −4.403 | 0.336 | −1.99 | NO | 482 | 4 |
| Cannabinol C2 | 268.4 | 2 | 1 | 93.96 | 0 | −4.834 | 0.5 | −1.32 | NO | 1310 | 4 |
| AChE | BChE | |||||||
|---|---|---|---|---|---|---|---|---|
| Compounds | B.A. (kcal/mol) | H bonds | Ki (μM) | Interactive Amino Acids | B.A. (kcal/mol) | H bonds | Ki (μM) | Interactive Amino Acids |
| THCV | −9.4 | 1 | 1.50 | VAL340, TYR337, TRP439, TYR449, GLY82, THR83, TRP86, VAL132, GLY121, TYR124, TRP286 | −8.9 | 0 | 3.25 | TYR440, VAL127, TRP82, GLY78, ASN83, SER79, PHE329, PRO285, PHE357, TYR332, SER72, PHE73, ASP70 |
| Cannabinol C2 | −9.3 | 1 | 4.42 | VAL340, TYR337, TRP439, TYR449, GLY82, THR83, TRP86, VAL132, GLY121, TYR124, TRP286 | −9.2 | 3 | 4.32 | TYR440, VAL127, GLU197, ALA202, ALA229, ASN397, TRP231, ALA199, SER198, GLU325, HIS438, TRP82, GLY78, SER79, PHE329, TYR332, GLY326, LEU286, THR204, ARG242, VAL228 |
| Cannabidiorcol | −9.4 | 2 | 4.61 | VAL340, TYR337, TRP439, TYR449, GLY82, THR83, TRP86, VAL132, GLY121, TRP286, TYR124 | −9.2 | 1 | 3.26 | GLY78, GLY435, ASN322, TYR440, VAL127, ARG424, MET437, LEU428, MET434, THR327, LYS339, VAL331, TRP430, ALA328, PHE76, TRP82 |
| Memantine | - | - | - | - | −6.8 | 1 | 0.54 | GLU197, TYR440, VAL127, TRP82, GLY78, SER79, VAL331, TYR332, PHE329, THR327, ALA328, GLY326, LEU286, GLU125, SER198, HIS438, MET437 |
| Donepezil | −8.4 | 2 | 1.46 | TYR337, TRP439, TYR449, VAL340, GLY82, THR83, GLY121, TRP86, VAL132, TRP286, TYR124 | - | - | - | - |
| Rivastigmine | −7.0 | 2 | 0.02 | TYR449, TRP439, TYR337, VAL240, GLY482, THR83, VAL132, TRP86, GLY121, TRP286, TYR124 | - | - | - | - |
| Galantamine | −7.1 | 0 | 2.1 | TYR337, TYR449, VAL340, TRP439, GLY82, THR83, TRP86, TYR124, GLY121, VAL132 | - | - | - | - |
| Compounds | AChE | BChE | ||||
|---|---|---|---|---|---|---|
| TE | VDW | HB | TE | VDW | HB | |
| THCV | −91.33 | −87.579 | −4.5 | −83.95 | −78.95 | −5 |
| Cannabinol C2 | −96.44 | −93.67 | 5 | −81.229 | −81.229 | 0 |
| Cannabidiorcol | −89.76 | −86.579 | −4.5 | −78.94 | −74.3765 | −4.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, N.; Chandel, V.; Rana, A.; Jain, M.; Kaushik, P. Investigation of Cannabis sativa Phytochemicals as Anti-Alzheimer’s Agents: An In Silico Study. Plants 2023, 12, 510. https://doi.org/10.3390/plants12030510
Patil N, Chandel V, Rana A, Jain M, Kaushik P. Investigation of Cannabis sativa Phytochemicals as Anti-Alzheimer’s Agents: An In Silico Study. Plants. 2023; 12(3):510. https://doi.org/10.3390/plants12030510
Chicago/Turabian StylePatil, Nil, Vaishnavi Chandel, Aarzu Rana, Mukul Jain, and Prashant Kaushik. 2023. "Investigation of Cannabis sativa Phytochemicals as Anti-Alzheimer’s Agents: An In Silico Study" Plants 12, no. 3: 510. https://doi.org/10.3390/plants12030510





