Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation
Abstract
1. Introduction
2. Results
2.1. Differences in Light Intensity and Flavonoid Accumulation
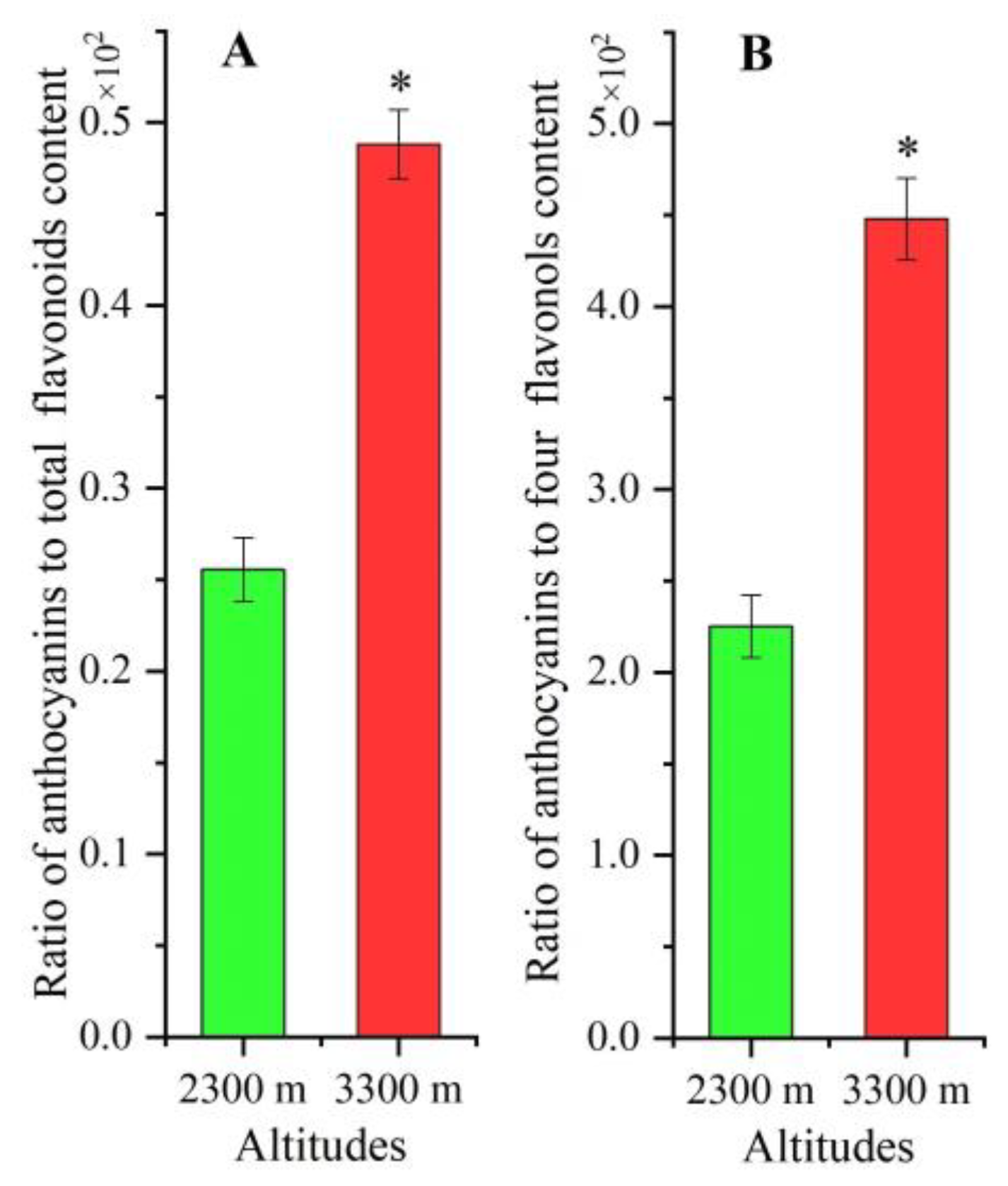
2.2. Ratio Differences for Anthocyanins and Total Flavonoids
2.3. Differentially Expressed Proteins at Higher Elevation
2.4. Differentially Expressed Genes Based on Elevation Differences
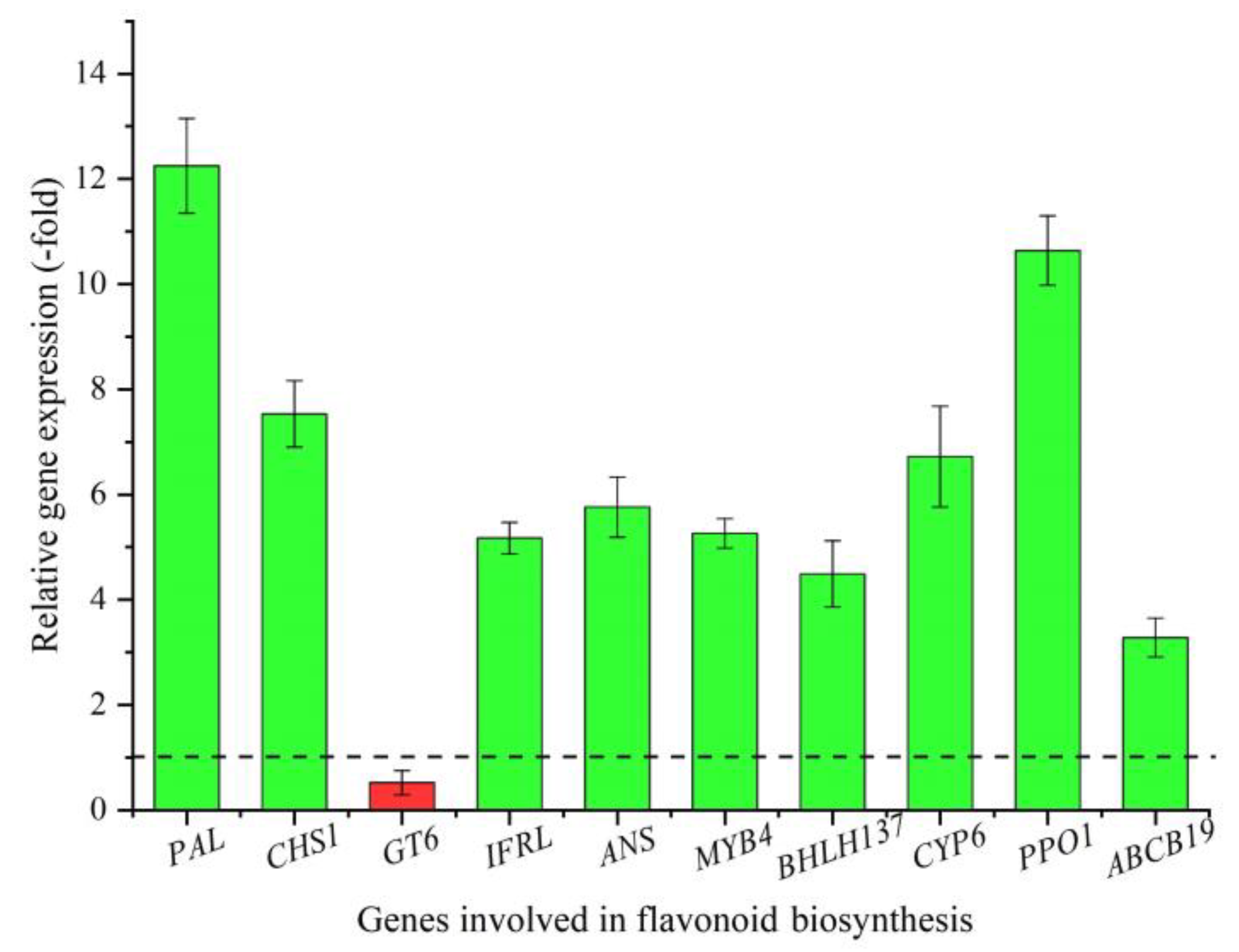
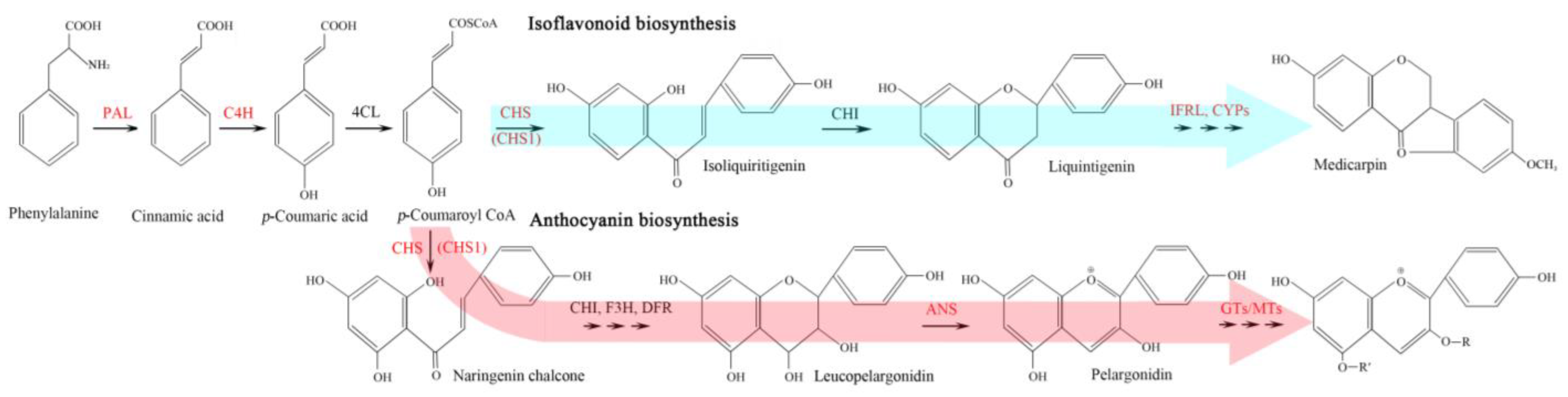
2.5. Expression Level of Genes Involved in Flavonoid Biosynthesis
2.6. Expression Level of Genes Involved in Light Stress
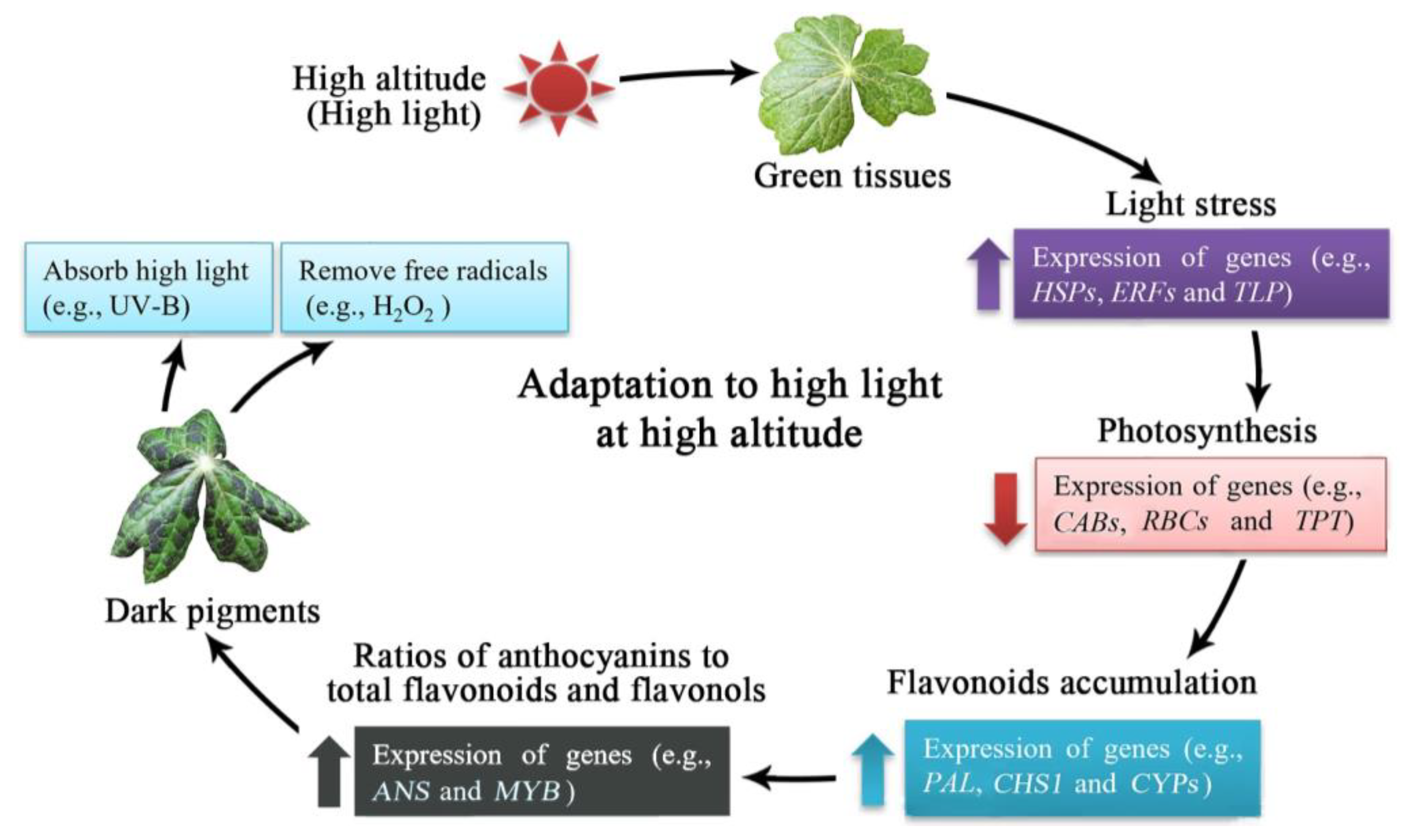
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Total Flavonoid, Flavonol, and Anthocyanin Contents
4.2.1. Extract Preparation
4.2.2. Determination of Total Flavonoid Content
4.2.3. Flavonol Quantification
4.2.4. Determination of Anthocyanin Content
4.3. Transcriptomic Analysis
4.4. Proteomic Analysis
4.4.1. Protein Extraction
4.4.2. 2-DE Separation
4.4.3. Gel Scanning and Image Analysis
4.4.4. Protein Identification
4.5. qRT-PCR Validation of Genes Involved in Light Stress and Flavonoids Biosynthesis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alam, M.A.; Naik, P.K. Impact of soil nutrients and environmental factors on podophyllotoxin content among 28 Podophyllum hexandrum populations of northwestern Himalayan region using linear and nonlinear. Commun. Soil Sci. Plant Anal. 2009, 40, 2485–2504. [Google Scholar] [CrossRef]
- Wu, A.L.; Li, M.; Zhang, S.W.; Zhao, J.F.; Liu, X.; Wang, C.H.; Wang, X.Y.; Zhong, G.Y. Effect of topographical factors on podophyllotoxin content in Sinopodophyllum hexandrum and study on ecological suitability. China J. Chin. Mater. Med. 2015, 40, 2299–2303. [Google Scholar]
- Li, M.F.; Ge, L.; Kang, T.L.; Sun, P.; Xing, H.; Yang, D.L.; Zhang, J.L.; Paré, P.W. High-elevation cultivation increases anti-cancer podophyllotoxin accumulation in Podophyllum hexandrum. Ind. Crops Prod. 2018, 121, 338–344. [Google Scholar] [CrossRef]
- Wang, Q.H.; Guo, S.; Yang, X.Y.; Zhang, Y.F.; Shang, M.Y.; Shang, Y.H.; Xiao, J.J.; Cai, S.Q. Flavonoids isolated from Sinopodophylli fructus and their bioactivities against human breast cancer cells. Chin. J. Nat. Med. 2017, 15, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Xu, H. Recent advances in semisynthesis, biosynthesis, biological activities, mode of action, and structure-activity relationship of podophyllotoxins: An update (2008–2010). Mini Rev. Med. Chem. 2011, 11, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.Y.; Xu, L.S.; Li, P.; Xu, G.J.; Wang, Y.X.; Cai, S.Q. Study on pharmacodynamics of Chinese herbal drug Guijiu and its lignan. Chin. Tradit. Herb. Drugs 2002, 33, 722–724. [Google Scholar]
- Choudhary, D.K.; Kaul, B.L.; Khan, S. Cultivation and conservation of Podophyllum hexandrum. An overview. J. Med. Aromat. Plant Sci. 1998, 20, 1071–1073. [Google Scholar]
- Gupta, M.L.; Dutta, A. Stress-mediated adaptive response leading to genetic diversity and instability in metabolite contents of high medicinal value: An overview on Podophyllum hexandrum. Omics 2011, 15, 873–882. [Google Scholar] [CrossRef]
- Kharkwal, A.C.; Kushwaha, R.; Prakash, O.; Ogra, R.K.; Bhattacharya, A.; Nagar, P.K.; Ahuja, P.S. An efficient method of propagation of Podophyllum hexandrum: An endangered medicinal plant of the Western Himalayas under ex situ conditions. J. Nat. Med. 2008, 62, 211–216. [Google Scholar] [CrossRef]
- Cao, X.L.; Li, M.L.; Li, J.; Song, Y.X.; Zhang, X.N.; Yang, D.L.; Li, M.F.; Wei, J.H. Co-expression of hydrolase genes improves seed germination of Sinopodophyllum hexandrum. Ind. Crops Prod. 2021, 64, 113414. [Google Scholar] [CrossRef]
- Sreenivasulu, Y.; Chanda, S.K.; Ahuja, P.S. Endosperm delays seed germination in Podophyllum hexandrum Royle—An important medicinal herb. Seed Sci. Technol. 2009, 37, 10–16. [Google Scholar] [CrossRef]
- Nadeem, M.; Palni, L.M.S.; Kumar, A.; Nandi, S.K. Podophyllotoxin content, above and belowground biomass in relation to altitude in Podophyllum hexandrum populations from Kumaun region of the Indian central Himalaya. Planta Med. 2007, 73, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Li, W.; Yang, D.L.; Zhou, L.L.; Li, T.T.; Su, X.M. Relationship between podophyllotoxin accumulation and soil nutrients and the influence of Fe2+ and Mn2+ on podophyllotoxin biosynthesis in Podophyllum hexandrum tissue culture. Plant Physiol. Biochem. 2013, 71, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Yin, D.; Zhao, X. Influence of ecological factors on the production of active substances in the anti-cancer plant Sinopodophyllum hexandrum (Royle) T.S. Ying. PLoS ONE 2015, 10, e0122981. [Google Scholar] [CrossRef]
- Pandey, H.; Kumar, A.; Palni, L.M.S.; Nandi, S.K. Podophyllotoxin content in rhizome and root samples of Podophyllum hexandrum Royle populations from Indian Himalayan region. J. Med. Plants Res. 2015, 9, 320–325. [Google Scholar]
- Kumari, A.; Singh, H.R.; Jha, A.; Swarnkar, M.K.; Shankar, R.; Kumar, S. Transcriptome sequencing of rhizome tissue of Sinopodophyllum hexandrum at two temperatures. BMC Genom. 2014, 15, 871. [Google Scholar] [CrossRef]
- Yang, D.L.; Sun, P.; Li, M.F. Chilling temperature stimulates growth, gene overexpression and podophyllotoxin biosynthesis in Podophyllum hexandrum Royle. Plant Physiol. Biochem. 2016, 107, 197–203. [Google Scholar] [CrossRef]
- Li, M.F.; Lv, M.; Yang, D.L.; Wei, J.H.; Xing, H.; Paré, P.W. Temperature-regulated anatomical and gene-expression changes in Sinopodophyllum hexandrum seedlings. Ind. Crops Prod. 2020, 152, 112479. [Google Scholar] [CrossRef]
- Kumari, A.; Dogra, V.; Joshi, R.; Kumar, S. Stress-responsive cis-regulatory elements underline podophyllotoxin biosynthesis and better performance of Sinopodophyllum hexandrum under water deficit conditions. Front. Plant Sci. 2022, 12, 751846. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.Q.; Li, H.E.; Gao, C.; Yang, R. Leaf traits and photosynthetic characteristics of endangered Sinopodophyllum hexandrum (Royle) Ying under different light regimes in Southeastern Tibet Plateau. Photosynthetica 2019, 57, 548–555. [Google Scholar] [CrossRef]
- Lv, M.; Su, H.Y.; Li, M.L.; Yang, D.L.; Yao, R.Y.; Li, M.F.; Wei, J.H. Effect of UV-B radiation on growth, flavonoid and podophyllotoxin accumulation, and related gene expression in Sinopodophyllum hexandrum. Plant Biol. 2020, 23, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Liu, M.Y.; Ruan, J.Y. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, G.; Cao, F.; Zhu, C.C.; Wang, G.Y.; El-Kassaby, Y.A. Light intensity affects the growth and flavonol biosynthesis of Ginkgo (Ginkgo biloba L.). New For. 2014, 45, 765–776. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef]
- Koyama, K.; Ikeda, H.; Poudel, P.R.; Goto-Yamamoto, N. Light quality affects flavonoid biosynthesis in young berries of Cabernet Sauvignon grape. Phytochemistry 2012, 78, 54–64. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Chivkunova, O.B. Light-stress-induced pigment changes and evidence for anthocyanin photoprotection in apples. J. Photochem. Photobiol. B 2000, 55, 155–163. [Google Scholar] [CrossRef]
- Winkel, B.S.J. Metabolic channeling in plants. Annu. Rev. Plant Biol. 2004, 55, 85–107. [Google Scholar] [CrossRef]
- Kleindt, C.K.; Stracke, R.; Mehrtens, F.; Weisshaar, B. Expression analysis of flavonoid biosynthesis genes during Arabidopsis thaliana silique and seed development with a primary focus on the proanthocyanidin biosynthetic pathway. BMC Res. Notes 2010, 3, 255. [Google Scholar] [CrossRef]
- Ayabe, S.I.; Akashi, T. Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 2006, 5, 271–282. [Google Scholar] [CrossRef]
- Frangne, N.; Eggmann, T.; Koblischke, C.; Weissenbock, G.; Martinoia, E.; Klein, M. Flavone glucoside uptake into barley mesophyll and Arabidopsis cell culture vacuoles. Energization occurs by H+-Antiport and ATP-binding cassette-type mechanisms1. Plant Physiol. 2002, 128, 726–733. [Google Scholar] [CrossRef]
- Zhu, T.T.; Zhang, M.H.; Su, H.Y.; Li, M.L.; Wang, Y.Y.; Jin, L.; Li, M.F. Integrated metabolomic and transcriptomic analysis reveals differential mechanism of flavonoid biosynthesis in two cultivars of Angelica sinensis. Molecules 2022, 27, 306. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.B.; Hu, Z.H. Preliminary studies on the distribution pattern and ecological adaptation of Sinopodophyllum hexandrum (Royle) ying (Berberidaceae). J. Wuhan Bot. Res. 1996, 14, 47–54. [Google Scholar]
- Rajkumar, S.; Ahuja, P.S. Developmental adaptation of leaves in Podophyllum hexandrum for effective pollination and dispersal. Curr. Sci. India 2010, 99, 1518–1519. [Google Scholar]
- Li, M.F.; Sun, P.; Kang, T.L.; Xing, H.; Yang, D.L.; Zhang, J.L.; Paré, P.W. Mapping podophyllotoxin biosynthesis and growth-related transcripts with high elevation in Sinopodophyllum hexandrum. Ind. Crops Prod. 2018, 124, 510–518. [Google Scholar] [CrossRef]
- Tohge, T.; De Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Hidalgo-Shrestha, C.; Bonawitz, N.D.; Franke, R.B.; Chapple, C. Spatio-temporal control of phenylpropanoid biosynthesis by inducible complementation of a cinnamate 4-hydroxylase mutant. J. Exp. Bot. 2021, 72, 3061–3073. [Google Scholar] [CrossRef]
- Santín, O.; Moncalián, G. Loading of malonyl-CoA onto tandem acyl carrier protein domains of polyunsaturated fatty acid synthases. J. Biol. Chem. 2018, 293, 12491–12501. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Patel, S.; Pan, X.P.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A.R. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Ahmad, N.; Liu, J.Y.; Xu, T.; Noman, M.; Jameel, A.; Yao, N.; Dong, Y.Y.; Wang, N.; Li, X.W.; Wang, F.W.; et al. Overexpression of a novel cytochrome P450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Virador, V.M.; Reyes, G.J.P.; Blanco-Labra, A.; Mendiola-Olaya, E.; Smith, G.M.; Moreno, A.; Whitaker, J.R. Cloning, sequencing, purification, and crystal structure of Grenache (Vitis vinifera) polyphenol oxidase. J. Agric. Food Chem. 2010, 58, 1189–1201. [Google Scholar] [CrossRef]
- Paiva, N.L.; Edwards, R.; Sun, Y.; Hrazdina, G.; Dixon, R.A. Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol. Biol. 1991, 17, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Peeters, A.J.; Leon-Kloosterziel, K.M.; Koornneef, M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 2001, 13, 853–871. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB tran-scription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Guo, C.; Chen, J.; Wu, Q.; Wang, X.; Chen, H. Integrated network analyses identify MYB4R1 neofunctionalization in the UV-B adaptation of tartary buckwheat. Plant Commun. 2022, 3, 100414. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef]
- Griesser, M.; Vitzthum, F.; Fink, B.; Bellido, M.L.; Raasch, C.; Munoz-Blanco, J.; Schwab, W. Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria × ananassa) achene and receptacle. J. Exp. Bot. 2008, 59, 2611–2625. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids--biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Z.; Suo, Y.; Wang, J.; Zheng, L.; Wang, Y. Gene expression and flavonol biosynthesis are induced by ultraviolet-B and salt stresses in Reaumuria trigyna. Biol. Plant. 2017, 61, 246–254. [Google Scholar] [CrossRef]
- Kim, T.; Samraj, S.; Jiménez, J.; Gómez, C.; Liu, T.; Begcy, K. Genome-wide identification of heat shock factors and heat shock proteins in response to UV and high intensity light stress in lettuce. BMC Plant Biol. 2021, 21, 185. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant. Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Kittang, A.I.; Winge, P.; van Loon, J.J.W.A.; Bones, A.M.; Iversen, T.H. Adaptation response of Arabidopsis thaliana to random positioning. Adv. Space Res. 2013, 52, 1320–1331. [Google Scholar] [CrossRef]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.S.; Deng, X.W.; Callis, J. Genome analysis and functional characterization of the E2 and RING–type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef]
- Jeon, E.H.; Pak, J.H.; Kim, M.J.; Kim, H.J.; Shin, S.H.; Lee, J.H.; Kim, D.H.; Oh, J.S.; Oh, B.J.; Jung, H.W.; et al. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2012, 427, 309–314. [Google Scholar] [CrossRef]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene responsive element binding factors act as transcriptional activators or repressors of GCC box mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed]
- Muoki, R.C.; Paul, A.; Kaachra, A.; Kumar, S. Membrane localized thaumatin-like protein from tea (CsTLP) enhanced seed yield and the plant survival under drought stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 163, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Dubey, R.S. Ascorbate peroxidase from rice seedlings: Properties of enzyme isoforms, effects of stresses and protective roles of osmolytes. Plant Sci. 2004, 167, 541–550. [Google Scholar] [CrossRef]
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 10270–10275. [Google Scholar] [CrossRef]
- Liu, S.Y.; Zhang, X.Q.; Wang, H.Y.; Yuan, Z.X.; Lei, H.X. Simultaneous determination of five major flavonoids in Tetrastigma hemsleyanum by HPLC. Chin. J. Mod. Appl. Pharm. 2018, 35, 1878–1880. [Google Scholar]
- Meng, X.C.; Zhang, Y.J.; Wang, X.Q. Content changes of anthocyanin, reducing sugar and soluble protein during the flower development of Petunia hybrid. J. South China Norm. Univ. 2001, 2, 96–99. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNAseq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Méchin, V.; Damerval, C.; Zivy, M. Total protein extraction with TCA-acetone. Methods Mol. Biol. 2007, 355, 1–8. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Planchon, S.; Renaut, J.; Vankova, R.; Prášil, I.T. Proteome analysis of cold response in spring and winter wheat (Triticum aestivum) crowns reveals similarities in stress adaptation and differences in regulatory processes between the growth habits. J. Proteome Res. 2013, 18, 4830–4845. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef]



| Genes | Proteins | No. | Nr ID | Spot Ratio (3300 m vs. 2300 m) |
|---|---|---|---|---|
| Metabolism (6) | ||||
| BGLU | Beta-1,3-glucanase | 28 | gi|458601845 | 4.57 |
| ANS | Anthocyanin synthase | 3 | gi|575498405 | 2.51 |
| CYP6 | Cytochrome P450 | 25 | gi|930809490 | 2.40 |
| CYP71BE30 | CYP71BE30 | 31 | gi|441418858 | 6.09 |
| PPO1 | Polyphenol oxidase | 16 | gi|930809512 | 3.30 |
| CS | Corytuberine synthase | 14 | gi|745698910 | 2.44 |
| Photosynthesis and energy (10) | ||||
| ycf3 | Photosystem I assembly protein ycf3 | 9 | gi|1021077178 | 29.30 |
| rbc | Ribulose-1,5-bisphosphate carboxylase/oxygenase | 2 | gi|6513628 | 2.26 |
| rbcL | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 5 | gi|825715491 | 11.33 |
| petL | Cytochrome b6-f complex subunit 6 | 22 | gi|1021077124 | 45.76 |
| atpA | ATP synthase CF1 alpha subunit | 24 | gi|918020571 | 54.42 |
| atpB | ATP synthase CF1 beta subunit | 8 | gi|918020573 | 2.39 |
| atpE | ATP synthase CF1 epsilon subunit | 26 | gi|918020601 | 3.41 |
| atpA | ATP synthase subunit alpha | 17 | gi|728802595 | 2.92 |
| matK | Maturase K | 20 | gi|356463749 | 5.73 |
| infA | Translation initiation factor IF-1 | 23 | gi|1025806949 | 2.27 |
| Cell morphogenesis (2) | ||||
| SMCP | Structural maintenance of chromosomes protein 2 | 10 | gi|913341257 | 79.04 |
| ACT | Actin | 15 | gi|307147595 | 9.70 |
| Transcription (4) | ||||
| PI | PISTILLATA-like protein | 1 | gi|186909195 | 2.85 |
| AP3-1 | APETALA3-like protein 1 | 7 | gi|186909191 | 5.54 |
| MYB4 | R2R3-MYB transcription factor MYB4 | 32 | gi|383290957 | 2.49 |
| FUL | FUL-like protein | 18 | gi|371941952 | 7.18 |
| Translation (8) | ||||
| rpl16 | Ribosomal protein L16 | 13 | gi|1025806946 | 4.58 |
| rpl20 | Ribosomal protein L20 | 29 | gi|1021076983 | 4.33 |
| rpl22 | Ribosomal protein L22 | 19 | gi|728802645 | 6.95 |
| rps2 | Ribosomal protein S2 | 21 | gi|1025806873 | 3.47 |
| rps11 | Ribosomal protein S11 | 6 | gi|1025806951 | 3.99 |
| rps12 | Ribosomal protein S12 | 11 | gi|552546114 | 2.33 |
| rps14 | Ribosomal protein S14 | 4 | gi|918020607 | 2.77 |
| rps16 | Ribosomal protein S16 | 27 | gi|728802592 | 4.29 |
| Genes | Proteins | SwissProt ID | log2 FC (3300 vs. 2300 m) |
|---|---|---|---|
| PAL | Phenylalanine ammonia-lyase | A0A059XSS3 | 4.18 |
| C4H | Cinnamic acid hydroxylase | Q43054 | 1.39 |
| CHS1 | Chalcone synthase 1 | P48386 | −5.02 |
| GT6 | UDP-glucose flavonoid 3-O-glucosyltransferase 6 | Q9SZG1 | −1.94 |
| IFRL | Isoflavone reductase-like protein | E1U332 | 2.07 |
| BHLH137 | Transcription factor bHLH137 | Q93W88 | 1.72 |
| DTX41 | Protein DETOXIFICATION 41 | Q9LYT3 | −1.61 |
| ABCB19 | ABC transporter B family member 19 | Q9LJX0 | 1.39 |
| CYP6 | Cytochrome P450 | A0A0N9HQE5 | 3.57 |
| CYP719A23 | CYP719A23 | L7T8H2 | 1.87 |
| CYP71CU1 | Cytochrome P450 family 71 subfamily CU polypeptide 1 | A0A0N9HTU1 | 1.39 |
| CYP734A1 | Cytochrome P450 734A1 | O48786 | 2.64 |
| CYP80B1 | (S)-N-methylcoclaurine 3′-hydroxylase isozyme 1 | O64899 | 3.49 |
| CYP81D11 | Cytochrome P450 81D11 | Q9LHA1 | 3.67 |
| CYP82A2 | Cytochrome P450 82A2 | O81972 | 3.15 |
| CYP82D47 | Cytochrome P450 CYP82D47 | H2DH24 | 2.23 |
| CYP94B3 | Cytochrome P450 94B3 | Q9SMP5 | −1.79 |
| PPO1 | Polyphenol oxidase | A0A0N9HGV8 | 4.87 |
| At5g38780 | S-adenosylmethionine-dependent methyltransferase At5g38780 | Q9FKR0 | −2.26 |
| Csa_1G002880 | Protein-L-isoaspartate O-methyltransferase | A0A0A0LRC6 | 1.51 |
| HMT1 | Homocysteine S-methyltransferase 1 | A4ZGQ8 | −2.78 |
| GXM3 | Glucuronoxylan 4-O-methyltransferase 3 | Q9LQ32 | 1.98 |
| Genes | Proteins | SwissProt ID | log2FC (3300 vs. 2300 m) |
|---|---|---|---|
| Photosynthesis (9) | |||
| CAB2 | Chlorophyll a-b binding protein 2 | P0CJ48 | −2.85 |
| CAB3 | Chlorophyll a-b binding protein 3 | P09756 | −2.09 |
| CAB8 | Chlorophyll a-b binding protein 8 | P27490 | 9.20 |
| CAB21 | Chlorophyll a-b binding protein 21 | P27493 | 10.72 |
| RBCS1 | Ribulose bisphosphate carboxylase small chain 1 | P16032 | −1.73 |
| RBCS4 | Ribulose bisphosphate carboxylase small chain 4 | Q39746 | −2.08 |
| RCAB | Ribulose bisphosphate carboxylase/oxygenase activase | Q42450 | −2.40 |
| TPT | Triose phosphate/phosphate translocator | P49132 | −2.52 |
| PSBR | Photosystem II 10 kDa polypeptide | P06183 | 1.93 |
| Light stress (10) | |||
| HSP18.1 | 18.1 kDa class I heat shock protein | P27879 | 7.52 |
| HSP90-1 | Heat shock protein 90-1 | P27323 | 3.08 |
| HSP22.0 | 22.0 kDa class IV heat shock protein | P30236 | 2.28 |
| HSP70 | Heat shock 70 kDa protein | O93866 | 1.54 |
| UBC4 | Ubiquitin-conjugating enzyme E2 4 | P42748 | 1.67 |
| ERF5 | Ethylene-responsive transcription factor 5 | O80341 | 7.18 |
| ERF9 | Ethylene-responsive transcription factor 9 | Q9FE67 | 4.02 |
| TLP | Thaumatin-like protein | Q53MB8 | 9.28 |
| APX3 | L-ascorbate peroxidase 3 | Q42564 | 2.22 |
| EX2 | Protein EXECUTER 2 | Q657X6 | 1.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Dong, M.; Li, M.; Jin, L.; Paré, P.W. Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation. Plants 2023, 12, 575. https://doi.org/10.3390/plants12030575
Zhao Q, Dong M, Li M, Jin L, Paré PW. Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation. Plants. 2023; 12(3):575. https://doi.org/10.3390/plants12030575
Chicago/Turabian StyleZhao, Qiaozhu, Miaoyin Dong, Mengfei Li, Ling Jin, and Paul W. Paré. 2023. "Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation" Plants 12, no. 3: 575. https://doi.org/10.3390/plants12030575
APA StyleZhao, Q., Dong, M., Li, M., Jin, L., & Paré, P. W. (2023). Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation. Plants, 12(3), 575. https://doi.org/10.3390/plants12030575







