Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis
Abstract
1. Introduction
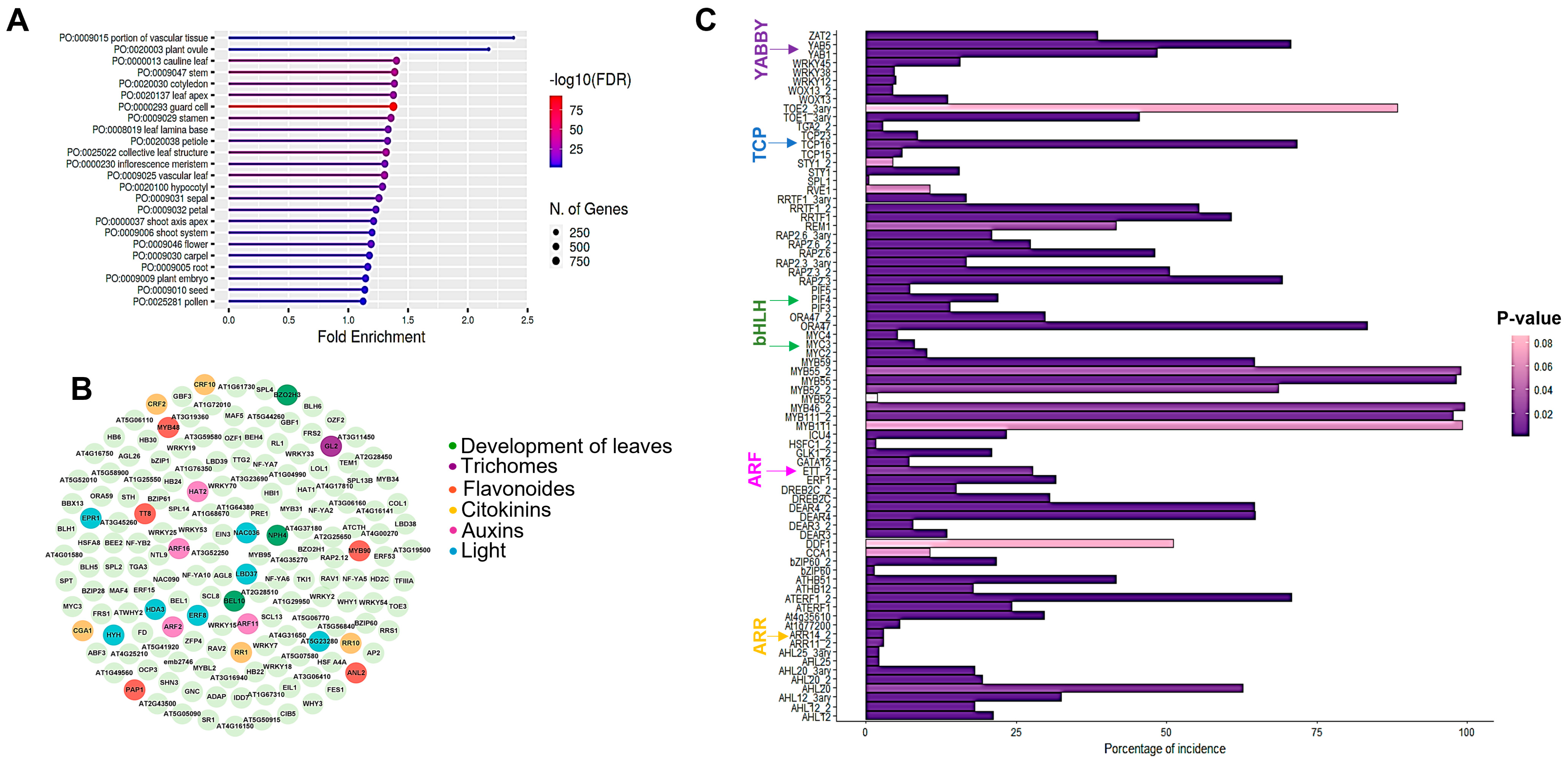
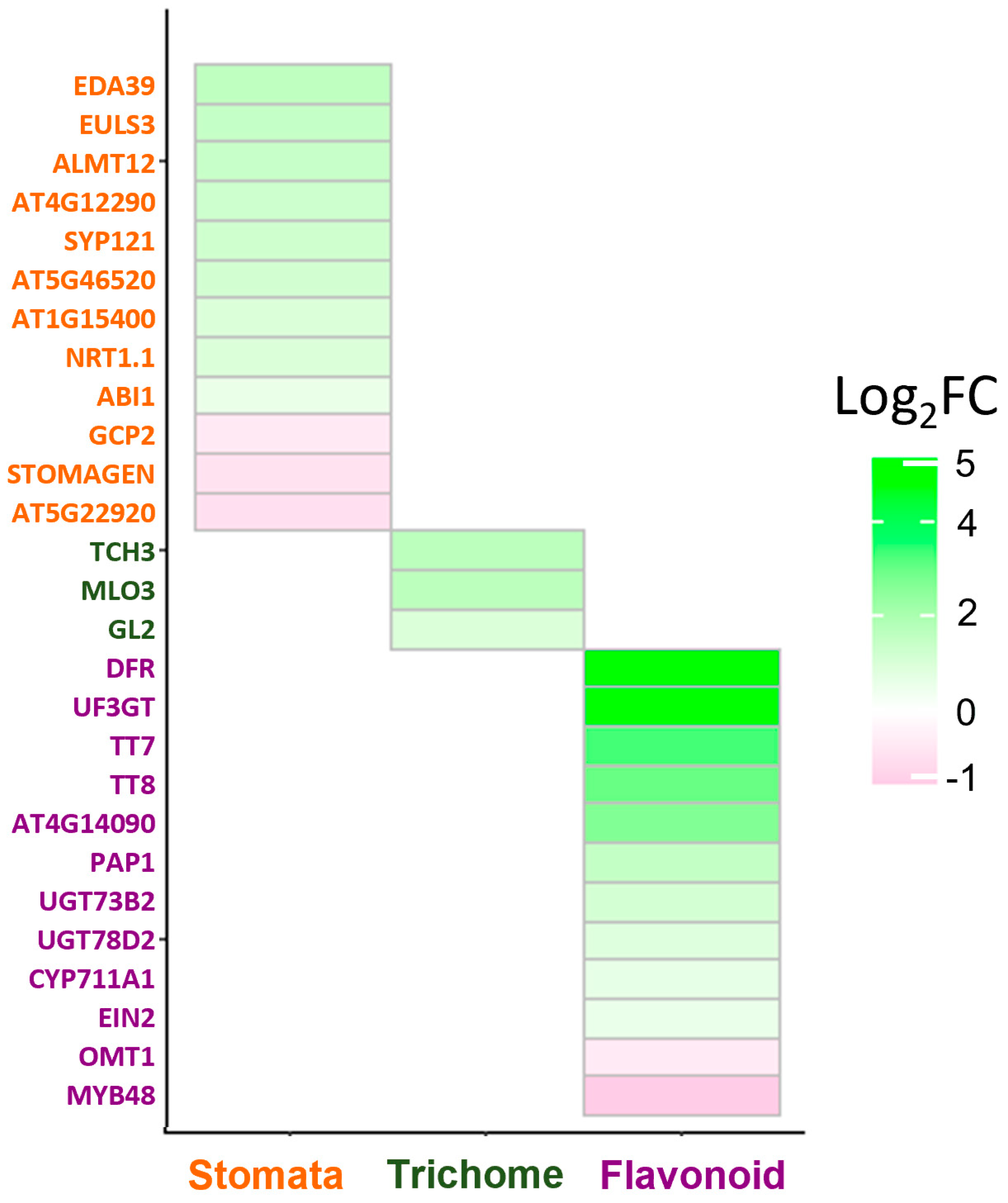
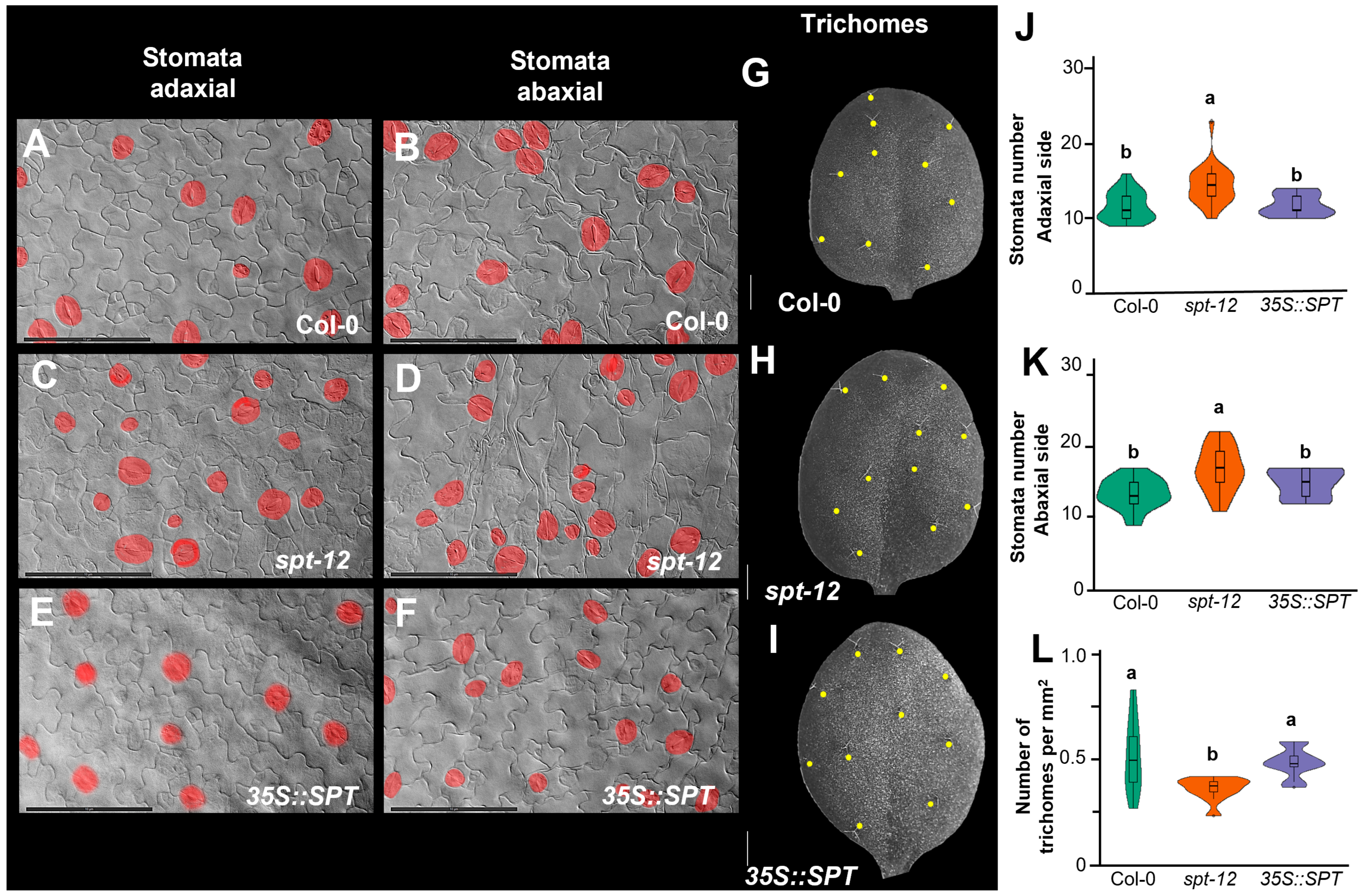
2. Results
2.1. SPT Contributes to the Control of Stomata and Trichome Number
2.2. SPT Is Required for the Control of Sucrose-Mediated Anthocyanin Induction
3. Discussion
4. Materials and Methods
4.1. Growth Conditions
4.2. RNA Extraction
4.3. RNA-Seq and Data Analyses
4.4. Aerial Measurement and Trichome Count
4.5. Microscopic Analysis of Stomata
4.6. Sucrose-Induced Anthocyanin Accumulation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, F.; Guan, C.; Jiao, Y. Molecular Mechanisms of Leaf Morphogenesis. Mol. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Vanhaeren, H.; Inzé, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef]
- Nikolov, L.A.; Runions, A.; Das Gupta, M.; Tsiantis, M. Leaf development and evolution. Curr. Top. Dev. Biol. 2019, 131, 109–139. [Google Scholar] [PubMed]
- Chen, L.; Wu, Z.; Hou, S. SPEECHLESS Speaks Loudly in Stomatal Development. Front. Plant Sci. 2020, 11, 114. [Google Scholar] [CrossRef]
- Le, J.; Zou, J.; Yang, K.; Wang, M.; Wang, S. Signaling to stomatal initiation and cell division. Front. Plant Sci. 2014, 5, 1–6. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Ohashi-Ito, K.; Bergmann, D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 2007, 445, 537–540. [Google Scholar] [CrossRef]
- De Marcos, A.; Houbaert, A.; Triviño, M.; Delgado, D.; Martín-Trillo, M.; Russinova, E.; Fenoll, C.; Mena, M. A Mutation in the bHLH Domain of the SPCH Transcription Factor Uncovers a BR-Dependent Mechanism for Stomatal Development. Plant Physiol. 2017, 174, 823–842. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505. [Google Scholar] [CrossRef]
- Pillitteri, L.J.; Bogenschutz, N.L.; Torii, K.U. The bHLH Protein, MUTE, Controls Differentiation of Stomata and the Hydathode Pore in Arabidopsis. Plant Cell Physiol. 2008, 49, 934–943. [Google Scholar] [CrossRef]
- Zoulias, N.; Harrison, E.L.; Casson, S.A.; Gray, J.E. Molecular control of stomatal development. Biochem. J. 2018, 475, 441–454. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Z.; Rahiman, R.; Lee, G.S.Y.; Yeo, Y.K.; Yang, X.; Lau, O.S. Light regulates stomatal development by modulating paracrine signaling from inner tissues. Nat. Commun. 2021, 12, 3403. [Google Scholar] [CrossRef] [PubMed]
- Schwab, B.; Folkers, U.; Hu, M. Trichome morphogenesis in Arabidopsis. R. Soc. 2000, 355, 879–883. [Google Scholar] [CrossRef]
- Fambrini, M.; Pugliesi, C. The Dynamic Genetic-Hormonal Regulatory Network Controlling the Trichome Development in Leaves. Plants 2019, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin Biosynthesis Induced by MYB Transcription Factors in Plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023, 361, 12–29. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Gusmaroli, G.; Allegra, D.; Galbiati, M.; Wade, H.K.; Jenkins, G.I.; Tonelli, C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 886–894. [Google Scholar] [CrossRef]
- Chen, S.; Wang, S. GLABRA2, a Common Regulator for Epidermal Cell Fate Determination and Anthocyanin Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4997. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Horiguchi, G.; Gleissberg, S.; Tsukaya, H. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 252–261. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef]
- Sidaway-Lee, K.; Josse, E.M.; Brown, A.; Gan, Y.; Halliday, K.J.; Graham, I.A.; Penfield, S. SPATULA links daytime temperature and plant growth rate. Curr. Biol. 2010, 20, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Vaistij, F.E.; Gan, Y.; Penfield, S.; Gilday, A.D.; Dave, A.; He, Z.; Josse, E.-M.; Choi, G.; Halliday, K.J.; Graham, I.A. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci. USA 2013, 110, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Upreti, S.; Yan, A.; Wakeel, A.; Wu, J.; Ge, S.; Liu, Y.; Liu, B.; Gan, Y. SPATULA regulates floral transition and photomorphogenesis in a PHYTOCHROME B-dependent manner in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 503, 2380–2385. [Google Scholar] [CrossRef] [PubMed]
- Reymond, M.C.; Brunoud, G.; Chauvet, A.; Martínez-Garcia, J.F.; Martin-Magniette, M.-L.; Monéger, F.; Scutt, C.P. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 2012, 24, 2812–2825. [Google Scholar] [CrossRef] [PubMed]
- Girin, T.; Paicu, T.; Stephenson, P.; Fuentes, S.; Korner, E.; O’Brien, M.; Sorefan, K.; Wood, T.A.; Balanzá, V.; Ferrándiz, C.; et al. INDEHISCENT and SPATULA Interact to Specify Carpel and Valve Margin Tissue and Thus Promote Seed Dispersal in Arabidopsis. Plant Cell 2011, 23, 3641–3653. [Google Scholar] [CrossRef]
- Groszmann, M.; Paicu, T.; Smyth, D.R. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 2008, 55, 40–52. [Google Scholar] [CrossRef]
- Groszmann, M.; Bylstra, Y.; Lampugnani, E.R.; Smyth, D.R. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 2010, 61, 1495–1508. [Google Scholar] [CrossRef]
- Groszmann, M.; Paicu, T.; Alvarez, J.P.; Swain, S.M.; Smyth, D.R. SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 2011, 68, 816–829. [Google Scholar] [CrossRef]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef]
- Nahar, M.A.U.; Ishida, T.; Smyth, D.R.; Tasaka, M.; Aida, M. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.M.; Kannangara, R.; Gilday, A.D.; Halliday, K.J.; Graham, I.A. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 2005, 15, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J.; Chavez Montes, R.A.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Gonzalez-Aguilera, K.L.; Ballester, P.; Ripoll, J.J.; Ezquer, I.; et al. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef] [PubMed]
- Franco-zorrilla, J.M.; López-vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Avramenko, T.V.; Sh Tsitsiashvili, G. Critical analysis of protein signaling networks involved in the regulation of plant secondary metabolism: Focus on anthocyanins. Crit. Rev. Biotechnol. 2016, 37, 685–700. [Google Scholar] [CrossRef]
- Herrera-Ubaldo, H.; Campos, S.E.; López-Gómez, P.; Luna-García, V.; Zúñiga-Mayo, V.M.; Armas-Caballero, G.E.; González-Aguilera, K.L.; DeLuna, A.; Marsch-Martínez, N.; Espinosa-Soto, C.; et al. The protein-protein interaction landscape of transcription factors during gynoecium development in Arabidopsis. Mol. Plant 2022, 16, 260–278. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Cherenkov, P.; Novikova, D.; Omelyanchuk, N.; Levitsky, V.; Grosse, I.; Weijers, D.; Mironova, V. Diversity of cis -regulatory elements associated with auxin response in Arabidopsis thaliana. J. Exp. Bot. 2018, 69, 329–339. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Lin, Y.; Hou, X. Ectopic Expression of a Pak-choi YABBY Gene, BcYAB3, Causes Leaf Curvature and Flowering Stage Delay in Arabidopsis thaliana. Genes 2020, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- She, Z.; Huang, X.; Aslam, M.; Wang, L.; Yan, M.; Qin, R.; Chen, Y.; Qin, Y.; Niu, X. Expression characterization and cross-species complementation uncover the functional conservation of YABBY genes for leaf abaxial polarity and carpel polarity establishment in Saccharum spontaneum. BMC Plant Biol. 2022, 22, 124. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yang, Z.P.; Zhang, J.; Zhang, L.G. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 2013, 14, 14872–14891. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martlnez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar]
- Hur, Y.S.; Kim, J.; Kim, S.; Son, O.; Kim, W.Y.; Kim, G.T.; Ohme-Takagi , M.; Cheon, C.-I. Identification of TCP13 as an Upstream Regulator of ATHB12 during Leaf Development. Genes 2019, 10, 644. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, H.; Zhang, C.; Li, X.; Lyu, Y.; Qi, D.; Cui, Y.; Hu, L.; Wang, Z.; Liang, Z.; et al. TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis. J. Integr. Plant Biol. 2019, 61, 1151–1170. [Google Scholar] [CrossRef] [PubMed]
- Kalve, S.; De Vos, D.; Beemster, G.T.S. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef]
- Skalák, J.; Vercruyssen, L.; Claeys, H.; Hradilová, J.; Černý, M.; Novák, O.; Plačková, L.; Saiz-Fernández, I.; Skaláková, P.; Coppens, F.; et al. Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. Plant J. 2019, 97, 805–824. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, A.; Zhou, Z.; Zhao, Y.; Yan, A.; Bao, S.; Yu, H.; Gan, Y. GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 2015, 206, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef] [PubMed]
- Vatén, A.; Soyars, C.L.; Tarr, P.T.; Nimchuk, Z.L.; Bergmann, D.C. Modulation of Asymmetric Division Diversity through Cytokinin and SPEECHLESS Regulatory Interactions in the Arabidopsis Stomatal Lineage. Dev. Cell 2018, 47, 53–66. [Google Scholar] [CrossRef]
- Xiong, Y.; Jiao, Y. The Diverse Roles of Auxin in Regulating Leaf Development. Plants 2019, 8, 243. [Google Scholar] [CrossRef]
- Zhang, J.Y.; He, S.B.; Li, L.; Yang, H.Q. Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc. Natl. Acad. Sci. USA 2014, 111, E3015–E3023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Runions, A.; Mentink, R.A.; Kierzkowski, D.; Karady, M.; Hashemi, B.; Huijser, P.; Strauss, S.; Gan, X.; Ljung, K.; et al. A WOX/Auxin Biosynthesis Module Controls Growth to Shape Leaf Form. Curr. Biol. 2020, 30, 4857–4868. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, L.; Zhao, Y.; An, L.; Yan, A.; Meng, X.; Gan, Y. Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol. 2013, 198, 699–708. [Google Scholar] [CrossRef]
- Hronková, M.; Wiesnerová, D.; Šimková, M.; Skůpa, P.; Dewitte, W.; Vráblová, M.; Zažímalová, E.; Šantrůček, J. Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J. Exp. Bot. 2015, 66, 4621–4630. [Google Scholar] [CrossRef]
- Sugano, S.S.; Shimada, T.; Imai, Y.; Okawa, K.; Tamai, A.; Mori, M.; Hara-Nishimura, I. Stomagen positively regulates stomatal density in Arabidopsis. Nature 2010, 463, 241–244. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana Flavonoid 3′-hydroxylase Gene and Functional Expression of the Encoded P450 Enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Messner, B.; Faus-Kessler, T.; Hoffmann, T.; Schwab, W.; Hajirezaei, M.R.; von Saint Paul, V.; Heller, W.; Schäffner, A.R. Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J. Exp. Bot. 2012, 63, 2465–2478. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.-I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Su, Y.S.; Lagarias, J.C. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol. Plant. 2009, 2, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Hu, Q.; Dai, X.; Tian, H.; Zheng, K.; Wang, X.; Mao, T.; Chen, J.-G.; Wang, S. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015, 83, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Bailey, T.L.; Grant, C.E. SEA: Simple Enrichment Analysis of motifs. BioRxiv 2021. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernal-Gallardo, J.J.; Zuñiga-Mayo, V.M.; Marsch-Martinez, N.; de Folter, S. Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis. Plants 2023, 12, 596. https://doi.org/10.3390/plants12030596
Bernal-Gallardo JJ, Zuñiga-Mayo VM, Marsch-Martinez N, de Folter S. Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis. Plants. 2023; 12(3):596. https://doi.org/10.3390/plants12030596
Chicago/Turabian StyleBernal-Gallardo, Judith Jazmin, Victor M. Zuñiga-Mayo, Nayelli Marsch-Martinez, and Stefan de Folter. 2023. "Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis" Plants 12, no. 3: 596. https://doi.org/10.3390/plants12030596
APA StyleBernal-Gallardo, J. J., Zuñiga-Mayo, V. M., Marsch-Martinez, N., & de Folter, S. (2023). Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis. Plants, 12(3), 596. https://doi.org/10.3390/plants12030596





