Illumina RNA and SMRT Sequencing Reveals the Mechanism of Uptake and Transformation of Selenium Nanoparticles in Soybean Seedlings
Abstract
:1. Introduction
2. Results
2.1. Total Se Content and Se Species in Soybean
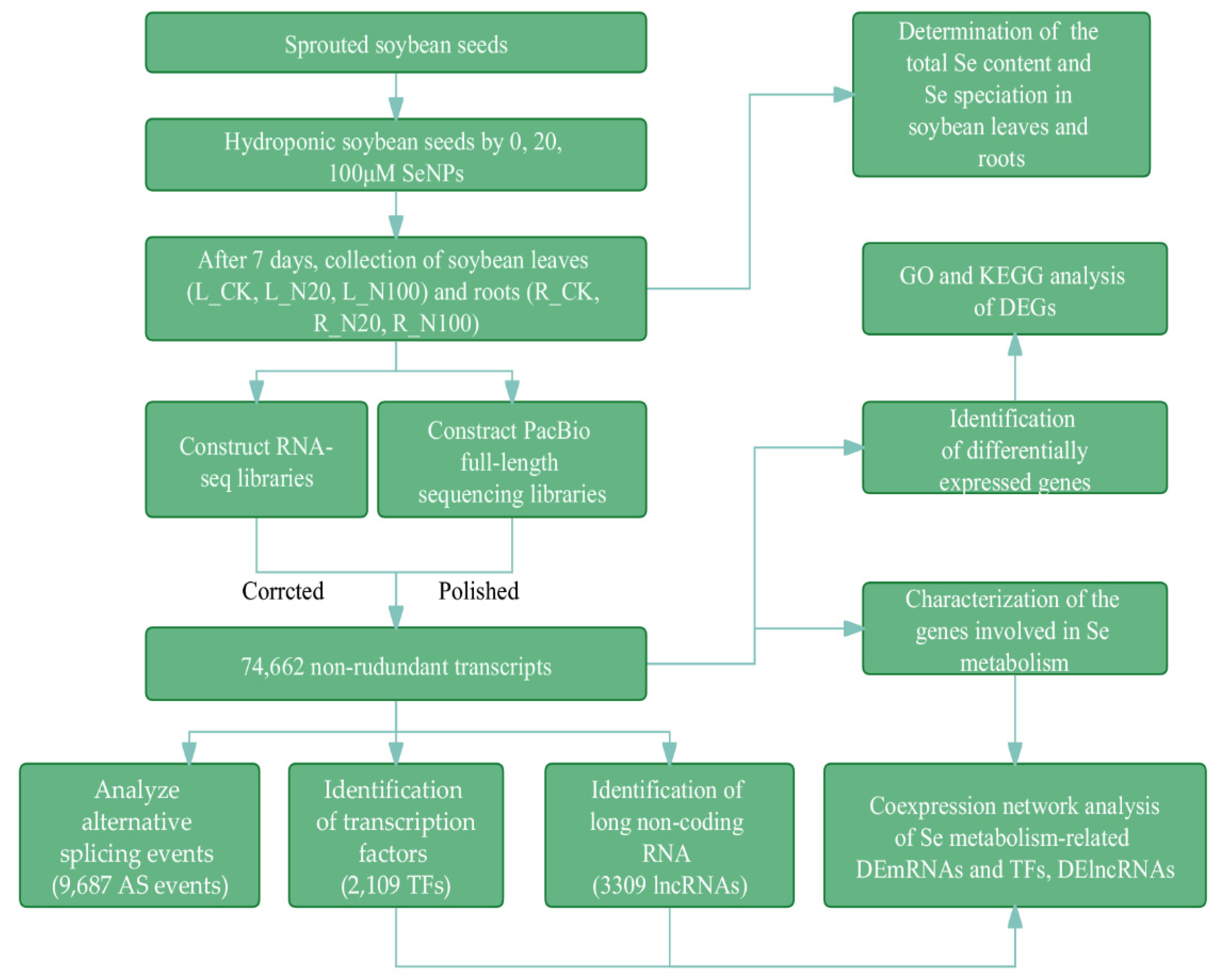
2.2. Sequencing Data Statistical Analysis of SMRT and Illumina RNA-Seq
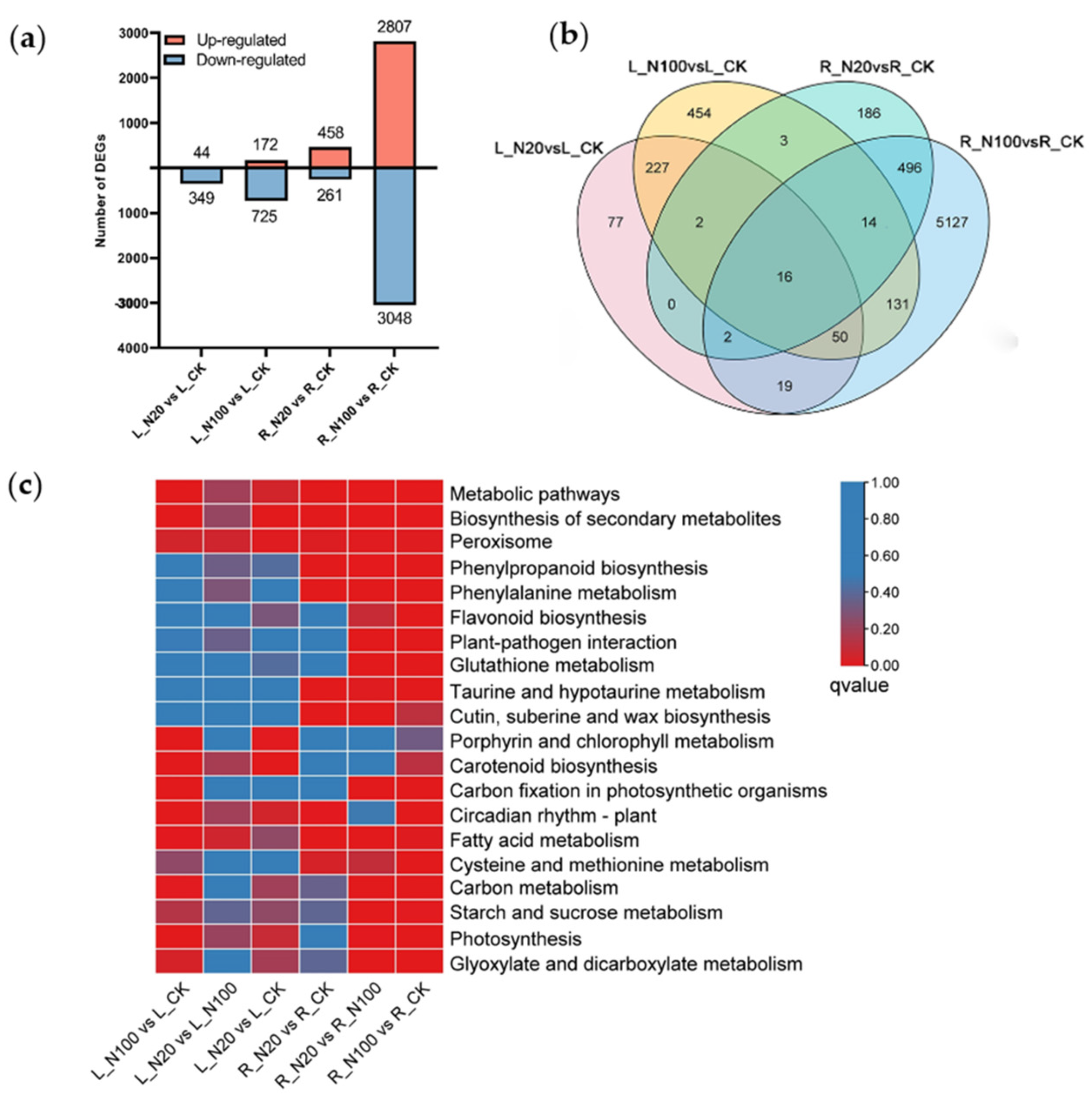
2.3. Analysis of DEGs in Response to Different SeNPs Treatment
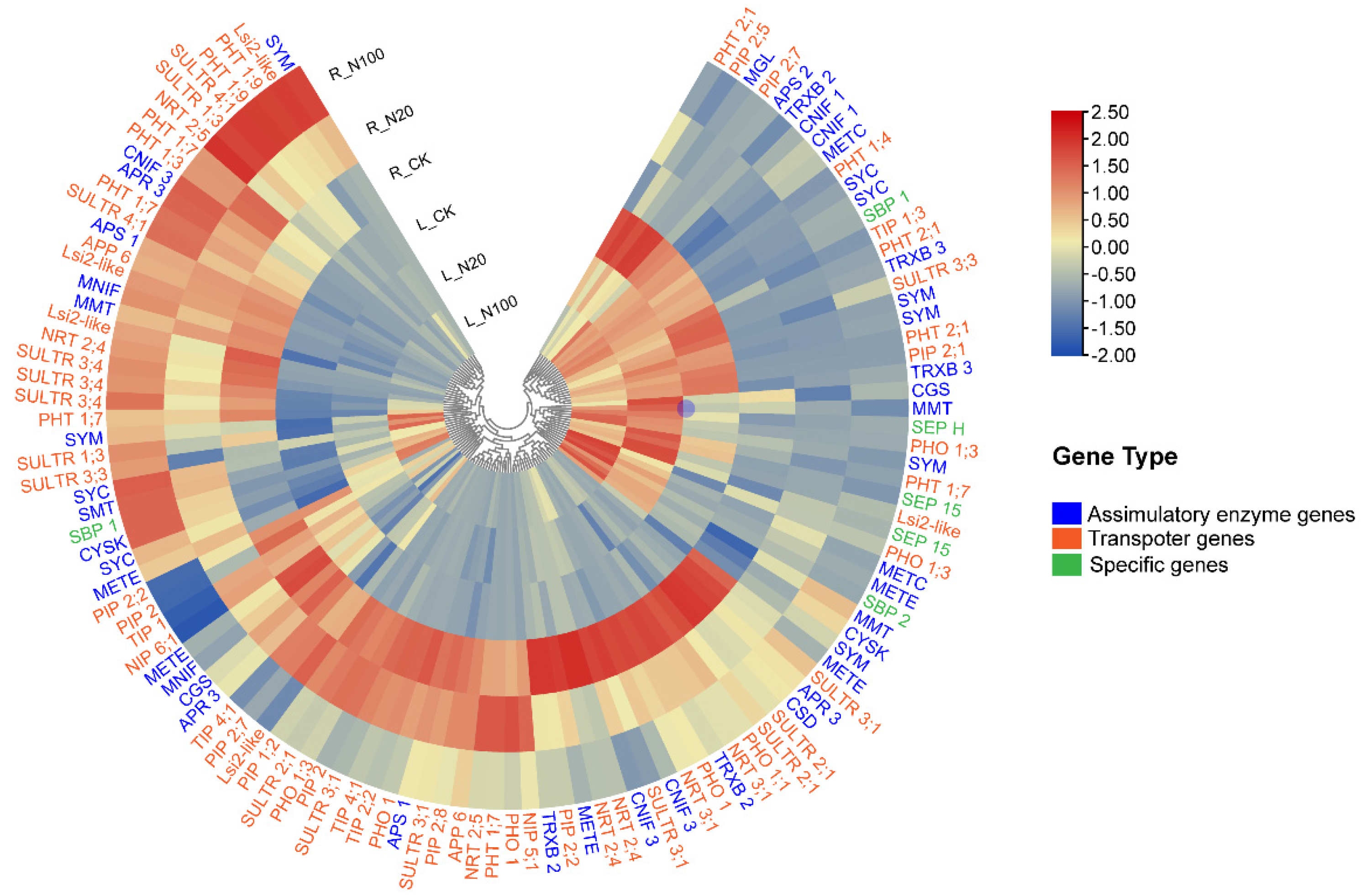
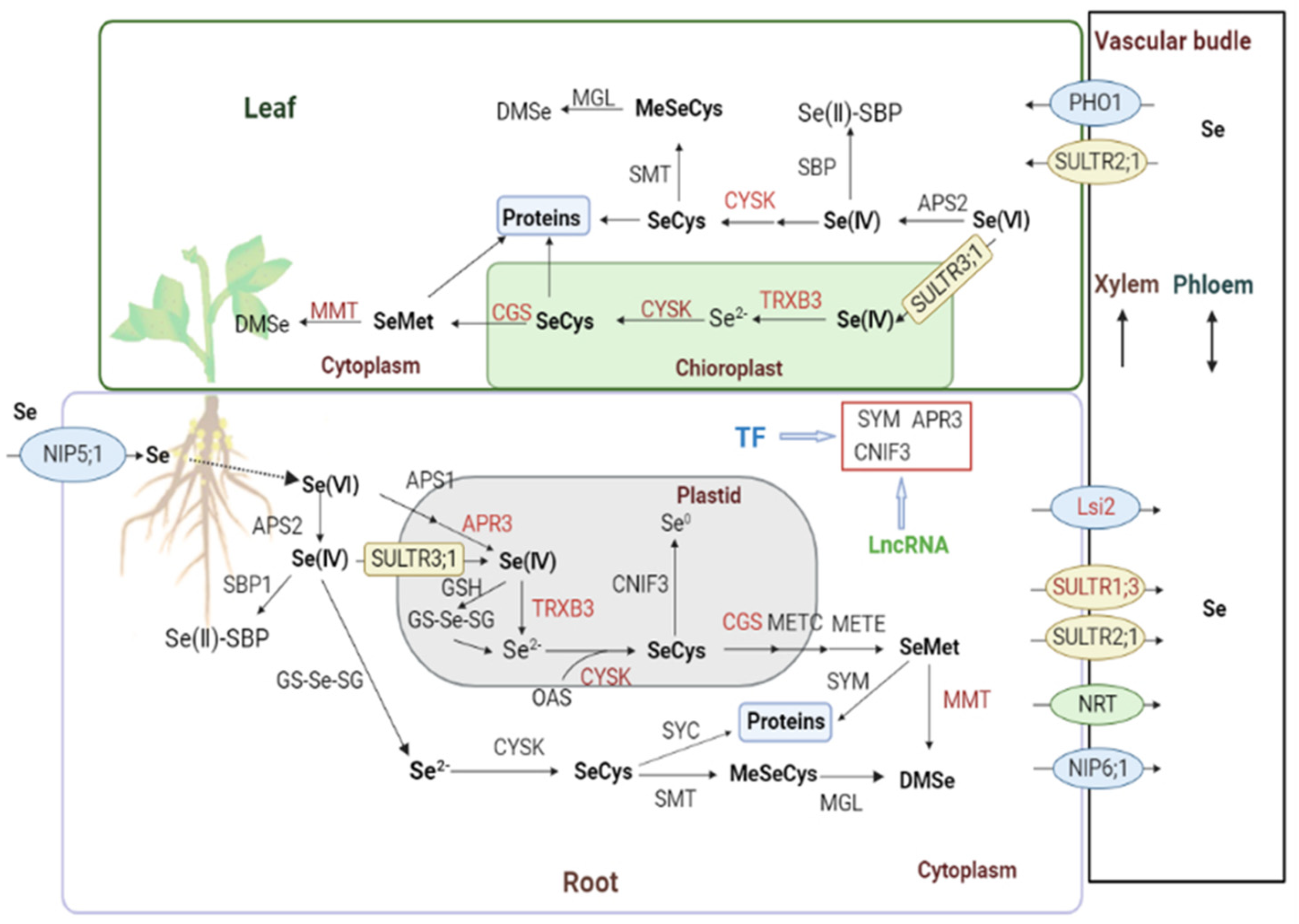
2.4. Characterization of the Genes Involved in Se Metabolism
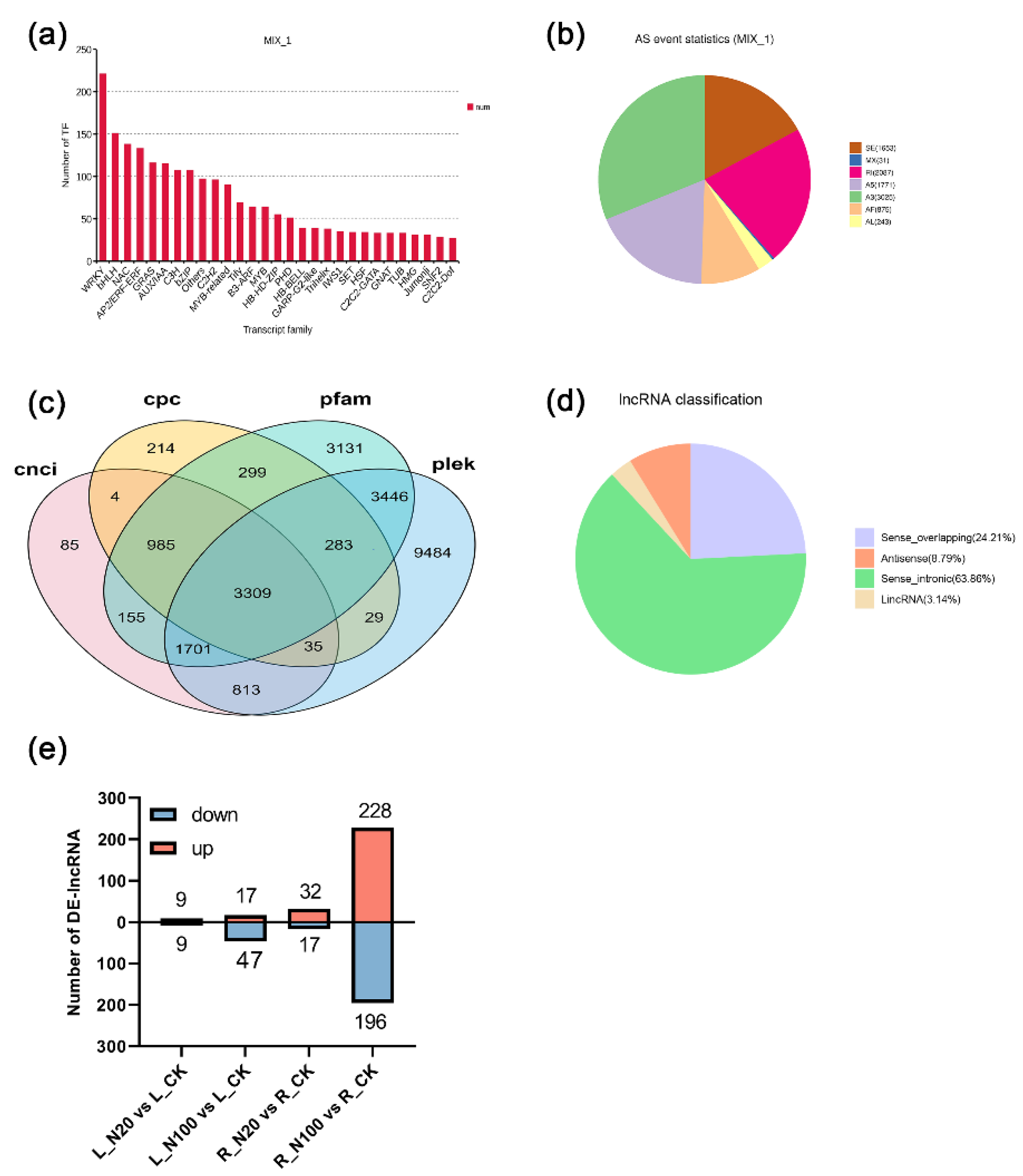
2.5. Identification of Transcription Factors (TFs), Alternative Splicing (AS) Events and lncRNAs
2.6. WGCNA Analysis of TFs and DE-lncRNAs Related to Se Metabolism in Soybean Supplied with SeNPs
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and SeNPs Treatment
4.2. Determination of Total Se Content and Se Species
4.3. RNA Isolation, Library Preparation, and Sequencing
4.4. Gene Functional Annotation and Identification of Transcripts Related to Se Metabolism
4.5. Identification of Transcription Factors, Alternative Splicing Events, and lncRNAs
4.6. Coexpression Network Analysis of Se Metabolism-Related DEmRNAs and TFs, DElncRNAs
4.7. Validation of RNA-seq Data by RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kieliszek, M. Selenium (-) Fascinating Microelement, Properties and Sources in Food. Molecules 2019, 24, 1298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, H.; Guo, Y.; Fan, B.; Huang, Y.; Mao, X.; Liang, K.; Hu, Z.; Sun, X.; Fang, Y.; et al. Benefit-risk assessment of dietary selenium and its associated metals intake in China (2017–2019): Is current seleni-um-rich agro-food safe enough? J. Hazard. Mater. 2020, 398, 123224. [Google Scholar] [CrossRef] [PubMed]
- Winkel, L.H.; Johnson, C.A.; Lenz, M.; Grundl, T.; Leupin, O.X.; Amini, M.; Charlet, L. Environmental selenium research: From microscopic processes to global understanding. Environ. Sci. Technol. 2012, 46, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Combs, G.J. Selenium in global food systems. Br. J. Nutr. 2001, 85, 517–547. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US) Panel on Dietary Antioxidants and Related Compounds. Dietary Reference Intakes for Vitamin C, Vitamin E, Selenium, and Carotenoids; National Academies Press (US): Washington, DC, USA, 2000.
- Schiavon, M.; Nardi, S.; Dalla, V.F.; Ertani, A. Selenium biofortification in the 21(st) century: Status and challenges for healthy human nutrition. Plant Soil 2020, 453, 245–270. [Google Scholar] [CrossRef]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Sarkar, S.; Ahmad, Z.; Vemuri, H.; Garai, S.; Mondal, M.; Bhatt, R.; et al. Selenium Biofortification: Roles, Mechanisms, Responses and Prospects. Molecules 2021, 26, 881. [Google Scholar] [CrossRef]
- Williams, P.N.; Lombi, E.; Sun, G.X.; Scheckel, K.; Zhu, Y.G.; Feng, X.; Zhu, J.; Carey, A.M.; Adomako, E.; Lawgali, Y.; et al. Selenium characterization in the global rice supply chain. Environ. Sci. Technol. 2009, 43, 6024–6030. [Google Scholar] [CrossRef]
- Bodnar, M.; Szczyglowska, M.; Konieczka, P.; Namiesnik, J. Methods of Selenium Supplementation: Bioavailability and Determination of Selenium Compounds. Crit. Rev. Food Sci. Nutr. 2016, 56, 36–55. [Google Scholar] [CrossRef]
- Zhang, Y.; Gladyshev, V.N. Comparative genomics of trace elements: Emerging dynamic view of trace element utilization and function. Chem. Rev. 2009, 109, 4828–4861. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Raza, A.; Hawrylak-Nowak, B.; Matraszek-Gawron, R.; Nahar, K.; Fujita, M. Selenium Toxicity in Plants and Environment: Biogeochemistry and Remediation Possibilities. Plants 2020, 9, 1711. [Google Scholar] [CrossRef]
- Kolbert, Z.; Molni, R.I.; Szollosi, R.K.; Feigl, G.B.; Erdei, L.S.; Ördög, A. Nitro-Oxidative Stress Correlates with Se Tolerance of Astragalus Species. Plant Cell Physiol. 2018, 59, 1827–1843. [Google Scholar] [CrossRef]
- Pilon-Smits, E.A.; Quinn, C.F.; Tapken, W.; Malagoli, M.; Schiavon, M. Physiological functions of beneficial elements. Curr. Opin. Plant Biol. 2009, 12, 267–274. [Google Scholar] [CrossRef]
- Lanza, M.; Reis, A. Roles of selenium in mineral plant nutrition: ROS scavenging responses against abiotic stresses. Plant Physiol. Biochem. 2021, 164, 27–43. [Google Scholar] [CrossRef]
- Wang, M.; Ali, F.; Qi, M.; Peng, Q.; Wang, M.; Banuelos, G.S.; Miao, S.; Li, Z.; Dinh, Q.T.; Liang, D. Insights into uptake, accumulation, and subcellular distribution of selenium among eight wheat (Triticum aestivum L.) cultivars supplied with selenite and selenate. Ecotoxicol. Environ. Saf. 2021, 207, 111544. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, N.; Liang, X.; Zheng, L.; Zhang, C.; Li, Y.F.; Zhang, Z.; Gao, Y.; Zhao, J. A comparative study on the accumulation, translocation and transformation of selenite, selenate, and SeNPs in a hydroponic-plant system. Ecotoxicol. Environ. Saf. 2020, 189, 109955. [Google Scholar] [CrossRef]
- White, P.J. Selenium accumulation by plants. Ann. Bot. 2016, 117, 217–235. [Google Scholar] [CrossRef]
- Schiavon, M.; Pilon-Smits, E.A. The fascinating facets of plant selenium accumulation—Biochemistry, physi-ology, evolution and ecology. New Phytol. 2017, 213, 1582–1596. [Google Scholar] [CrossRef]
- Schiavon, M.; Pilon, M.; Malagoli, M.; Pilon-Smits, E.A. Exploring the importance of sulfate transporters and ATP sulphurylases for selenium hyperaccumulation-a comparison of Stanleya pinnata and Brassica juncea (Brassicaceae). Front. Plant Sci. 2015, 6, 2. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, B.; Li, W.; Che, R.; Deng, K.; Li, H.; Yu, F.; Ling, H.; Li, Y.; Chu, C. OsPT2, a phosphate transporter, is involved in the active uptake of selenite in rice. New Phytol. 2014, 201, 1183–1191. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Mitani, N.; Yamaji, N.; Shen, R.F.; Ma, J.F. Involvement of silicon influx transporter OsNIP2;1 in selenite uptake in rice. Plant Physiol. 2010, 153, 1871–1877. [Google Scholar] [CrossRef] [Green Version]
- De Souza, M.P.; Pilon-Smits, E.A.; Lytle, C.M.; Hwang, S.; Tai, J.; Honma, T.S.; Yeh, L.; Terry, N. Rate-limiting steps in selenium assimilation and volatilization by indian mustard. Plant Physiol. 1998, 117, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Kolbert, Z.; Molnar, A.; Feigl, G.; Van Hoewyk, D. Plant selenium toxicity: Proteome in the crosshairs. J. Plant Physiol. 2019, 232, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Neuhierl, B.; Bock, A. On the mechanism of selenium tolerance in selenium-accumulating plants. Purification and characterization of a specific selenocysteine methyltransferase from cultured cells of Astragalus bis-culatus. Eur. J. Biochem. 1996, 239, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Van Huysen, T.; Abdel-Ghany, S.; Hale, K.L.; LeDuc, D.; Terry, N.; Pilon-Smits, E.A. Overexpression of cysta-thionine-gamma-synthase enhances selenium volatilization in Brassica juncea. Planta 2003, 218, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Esaki, N.; Seraneeprakarn, V.; Tanaka, H.; Soda, K. Purification and characterization of Clostridium sticklandii D-selenocystine alpha, beta-lyase. J. Bacteriol. 1988, 170, 751–756. [Google Scholar] [CrossRef]
- Trippe, R.R.; Pilon-Smits, E. Selenium transport and metabolism in plants: Phytoremediation and biofortification implications. J. Hazard. Mater. 2021, 404, 124178. [Google Scholar] [CrossRef]
- Lima, L.W.; Pilon-Smits, E.; Schiavon, M. Mechanisms of selenium hyperaccumulation in plants: A survey of molecular, biochemical and ecological cues. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2343–2353. [Google Scholar] [CrossRef]
- Van Hoewyk, D.; Garifullina, G.F.; Ackley, A.R.; Abdel-Ghany, S.E.; Marcus, M.A.; Fakra, S.; Ishiyama, K.; Inoue, E.; Pilon, M.; Takahashi, H.; et al. Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiol. 2005, 139, 1518–1528. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, K.S. Role of nano-selenium in health and environment. J. Biotechnol. 2021, 325, 152–163. [Google Scholar] [CrossRef]
- Moreno-Martin, G.; Sanz-Landaluze, J.; Leon-Gonzalez, M.E.; Madrid, Y. Insights into the accumulation and transformation of Ch-SeNPs by Raphanus sativus and Brassica juncea: Effect on essential elements uptake. Sci. Total Environ. 2020, 725, 138453. [Google Scholar] [CrossRef]
- Li, D.; Zhou, C.; Zhang, J.; An, Q.; Wu, Y.; Li, J.Q.; Pan, C. Nanoselenium Foliar Applications Enhance the Nutrient Quality of Pepper by Activating the Capsaicinoid Synthetic Pathway. J. Agric. Food Chem. 2020, 68, 9888–9895. [Google Scholar] [CrossRef]
- Ferro, C.; Florindo, H.F.; Santos, H.A. Selenium Nanoparticles for Biomedical Applications: From Development and Characterization to Therapeutics. Adv. Healthc. Mater. 2021, 10, e2100598. [Google Scholar] [CrossRef]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef]
- Liu, J.; Qi, W.Y.; Chen, H.; Song, C.; Li, Q.; Wang, S.G. Selenium Nanoparticles as an Innovative Selenium Fer-tilizer Exert Less Disturbance to Soil Microorganisms. Front. Microbiol. 2021, 12, 746046. [Google Scholar] [CrossRef]
- Gallego-Gallegos, M.; Doig, L.E.; Tse, J.J.; Pickering, I.J.; Liber, K. Bioavailability, toxicity and biotransformation of selenium in midge (Chironomus dilutus) larvae exposed via water or diet to elemental selenium particles, selenite, or selenized algae. Environ. Sci. Technol. 2013, 47, 584–592. [Google Scholar] [CrossRef]
- Neysanian, M.; Iranbakhsh, A.; Ahmadvand, R.; Oraghi, A.Z.; Ebadi, M. Comparative efficacy of selenate and selenium nanoparticles for improving growth, productivity, fruit quality, and postharvest longevity through modifying nutrition, metabolism, and gene expression in tomato; potential benefits and risk assessment. PLoS ONE 2020, 15, e244207. [Google Scholar] [CrossRef]
- Sotoodehnia-Korani, S.; Iranbakhsh, A.; Ebadi, M.; Majd, A.; Oraghi, A.Z. Selenium nanoparticles induced variations in growth, morphology, anatomy, biochemistry, gene expression, and epigenetic DNA methylation in Capsicum annuum; an in vitro study. Environ. Pollut. 2020, 265, 114727. [Google Scholar] [CrossRef]
- Safari, M.; Oraghi Ardebili, Z.; Iranbakhsh, A. Selenium nano-particle induced alterations in expression patterns of heat shock factor A4A (HSFA4A), and high molecular weight glutenin subunit 1Bx (Glu-1Bx) and enhanced nitrate reductase activity in wheat (Triticum aestivum L.). Acta Physiol. Plant. 2018, 40, 117. [Google Scholar] [CrossRef]
- Escalante-Valdez, M.J.; Guardado-Felix, D.; Serna-Saldivar, S.O.; Barrera-Arellano, D.; Chuck-Hernandez, C. Effects of Post Anthesis Foliar Application of Sodium Selenite to Soybeans (Glycine max): Lipid Composition and Oil Stability. Biomolecules 2019, 9, 772. [Google Scholar] [CrossRef]
- Zhang, X.; He, H.; Xiang, J.; Li, B.; Zhao, M.; Hou, T. Selenium-containing soybean antioxidant peptides: Prep-aration and comprehensive comparison of different selenium supplements. Food Chem. 2021, 358, 129888. [Google Scholar] [CrossRef]
- Deng, X.; Liu, K.; Li, M.; Zhang, W.; Zhao, X.; Zhao, Z.; Liu, X. Difference of selenium uptake and distribution in the plant and selenium form in the grains of rice with foliar spray of selenite or selenate at different stages. Field Crop. Res. 2017, 211, 165–171. [Google Scholar] [CrossRef]
- Deng, X.; Liao, J.; Zhao, Z.; Qin, Y.; Liu, X. Distribution and speciation of selenium in soybean proteins and its effect on protein structure and functionality. Food Chem. 2022, 370, 130982. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Y.; Li, K.; Wan, Y.; Wang, Q.; Zhuang, Z.; Guo, Y.; Li, H. Uptake, translocation and biotrans-formation of selenium nanoparticles in rice seedlings (Oryza sativa L.). J. Nanobiotechnol. 2020, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Li, H.; Li, J.; Zhao, G.; Wu, W.; Liu, L.; Wang, Q.; Guo, Y. Absorption and Bio-Transformation of Selenium Nanoparticles by Wheat Seedlings (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Dou, L.; Tian, Z.; Zhao, Q.; Xu, M.; Zhu, Y.; Luo, X.; Qiao, X.; Ren, R.; Zhang, X.; Li, H. Transcriptomic Charac-terization of the Effects of Selenium on Maize Seedling Growth. Front. Plant Sci. 2021, 12, 2525. [Google Scholar] [CrossRef]
- Rao, S.; Xiao, X.; Wang, Y.; Xiong, Y.; Cheng, H.; Li, L.; Cheng, S. Comparative study of the effects of selenium nanoparticles and selenite on selenium content and nutrient quality in soybean sprouts. Folia Hortic. 2022, 34, 223–234. [Google Scholar] [CrossRef]
- Wang, M.; Peng, Q.; Zhou, F.; Yang, W.; Dinh, Q.T.; Liang, D. Uptake kinetics and interaction of selenium species in tomato (Solanum lycopersicum L.) seedlings. Environ. Sci. Pollut. Res. 2019, 26, 9730–9738. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Li, S.; Zhang, L.; Qi, C.; Weeda, S.; Zhao, B.; Ren, S.; Guo, Y.D. Plasma Membrane Intrinsic Proteins SlPIP2;1, SlPIP2;7 and SlPIP2;5 Conferring Enhanced Drought Stress Tolerance in Tomato. Sci. Rep. 2016, 6, 31814. [Google Scholar] [CrossRef]
- Yamaji, N.; Ma, J.F. Metalloid transporters and their regulation in plants. Plant Physiol. 2021, 187, 1929–1939. [Google Scholar] [CrossRef]
- Cao, M.J.; Wang, Z.; Wirtz, M.; Hell, R.; Oliver, D.J.; Xiang, C.B. SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 2013, 73, 607–616. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Inoue, E.; Saito, K.; Yamaya, T.; Takahashi, H. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 2003, 131, 1511–1517. [Google Scholar] [CrossRef]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef]
- Hamburger, D.; Rezzonico, E.; MacDonald-Comber, P.J.; Somerville, C.; Poirier, Y. Identification and charac-terization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, H.; He, L.; Zhu, W.; Yan, L.; Chen, Q.; He, C. The PHOSPHATE1 genes participate in salt and Pi signaling pathways and play adaptive roles during soybean evolution. BMC Plant Biol. 2019, 19, 353. [Google Scholar] [CrossRef]
- Cao, D.; Liu, Y.; Linlong, M.; Liu, Z.; Li, J.; Beibei, W.; Zhang, X.; Yin, P.; Jin, X.; Huang, J. Genome-wide identifi-cation and characterization of phosphate transporter gene family members in tea plants (Camellia sinensis L. O. kuntze) under different selenite levels. Plant Physiol. Biochem. 2021, 166, 668–676. [Google Scholar] [CrossRef]
- Yang, X.; Liao, X.; Yu, L.; Rao, S.; Chen, Q.; Zhu, Z.; Cong, X.; Zhang, W.; Ye, J.; Cheng, S.; et al. Combined metabolome and transcriptome analysis reveal the mechanism of selenate influence on the growth and quality of cabbage (Brassica oleracea var. capitata L.). Food Res. Int. 2022, 156, 111135. [Google Scholar] [CrossRef]
- Funes-Collado, V.; Morell-Garcia, A.; Rubio, R.; Lopez-Sanchez, J.F. Study of selenocompounds from sele-nium-enriched culture of edible sprouts. Food Chem. 2013, 141, 3738–3743. [Google Scholar] [CrossRef]
- Huang, Y.; Fan, B.; Lei, N.; Xiong, Y.; Liu, Y.; Tong, L.; Wang, F.; Maesen, P.; Blecker, C. Selenium Biofortification of Soybean Sprouts: Effects of Selenium Enrichment on Proteins, Protein Structure, and Functional Properties. Front. Nutr. 2022, 9, 849928. [Google Scholar] [CrossRef]
- Grant, K.; Carey, N.M.; Mendoza, M.; Schulze, J.; Pilon, M.; Pilon-Smits, E.A.; van Hoewyk, D. Adenosine 5′-phosphosulfate reductase (APR2) mutation in Arabidopsis implicates glutathione deficiency in selenate toxicity. Biochem. J. 2011, 438, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, A.; Tobe, R.; Aono, R.; Inoue, M.; Hagita, S.; Kiriyama, K.; Toyotake, Y.; Ogawa, T.; Kurihara, T.; Goto, K.; et al. Initial Step of Selenite Reduction via Thioredoxin for Bacterial Selenoprotein Biosynthesis. Int. J. Mol. Sci. 2021, 22, 10965. [Google Scholar] [CrossRef] [PubMed]
- Schild, F.; Kieffer-Jaquinod, S.; Palencia, A.; Cobessi, D.; Sarret, G.; Zubieta, C.; Jourdain, A.; Dumas, R.; Forge, V.; Testemale, D.; et al. Biochemical and biophysical characterization of the selenium-binding and reducing site in Arabidopsis thaliana homologue to mammals selenium-binding protein 1. J. Biol. Chem. 2014, 289, 31765–31776. [Google Scholar] [CrossRef] [PubMed]
- Fisher, B.; Yarmolinsky, D.; Abdel-Ghany, S.; Pilon, M.; Pilon-Smits, E.A.; Sagi, M.; Van Hoewyk, D. Superoxide generated from the glutathione-mediated reduction of selenite damages the iron-sulfur cluster of chloro-plastic ferredoxin. Plant Physiol. Biochem. 2016, 106, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Van Hoewyk, D. A tale of two toxicities: Malformed selenoproteins and oxidative stress both contribute to selenium stress in plants. Ann. Bot. 2013, 112, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Joshi, V.; Jander, G. The catabolic enzyme methionine gamma-lyase limits methionine accumulation in potato tubers. Plant Biotechnol. J. 2014, 12, 883–893. [Google Scholar] [CrossRef]
- Tagmount, A.; Berken, A.; Terry, N. An essential role of s-adenosyl-L-methionine:L-methionine s-methyltransferase in selenium volatilization by plants. Methylation of selenomethionine to seleni-um-methyl-L-selenium- methionine, the precursor of volatile selenium. Plant Physiol. 2002, 130, 847–856. [Google Scholar] [CrossRef]
- Liu, J.; Chen, S.; Liu, M.; Chen, Y.; Fan, W.; Lee, S.; Xiao, H.; Kudrna, D.; Li, Z.; Chen, X.; et al. Full-Length Tran-scriptome Sequencing Reveals Alternative Splicing and lncRNA Regulation during Nodule Development in Glycine max. Int. J. Mol. Sci. 2022, 23, 7371. [Google Scholar] [CrossRef]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Trivedi, P.K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genom. 2011, 11, 259–273. [Google Scholar] [CrossRef]
- Cui, G.; Chai, H.; Yin, H.; Yang, M.; Hu, G.; Guo, M.; Yi, R.; Zhang, P. Full-length transcriptome sequencing reveals the low-temperature-tolerance mechanism of Medicago falcata roots. BMC Plant Biol. 2019, 19, 575. [Google Scholar] [CrossRef]
- Rao, S.; Yu, T.; Cong, X.; Xu, F.; Lai, X.; Zhang, W.; Liao, Y.; Cheng, S. Integration analysis of PacBio SMRT- and Illumina RNA-seq reveals candidate genes and pathway involved in selenium metabolism in hyperaccu-mulator Cardamine violifolia. BMC Plant Biol. 2020, 20, 492. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera, A.R.; Landsman, D.; Koonin, E.V. COG database update: Focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein family’s database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Gerttula, S.; Zinkgraf, M.; Muday, G.K.; Lewis, D.R.; Ibatullin, F.M.; Brumer, H.; Hart, F.; Mansfield, S.D.; Filkov, V.; Groover, A. Transcriptional and Hormonal Regulation of Gravitropism of Woody Stems in Populus. Plant Cell 2015, 27, 2800–2813. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Zhang, Y.; Wang, X.; Yang, S.; Fang, T.; Zheng, S.; Zeng, L. Integrative mRNA and Long Noncoding RNA Analysis Reveals the Regulatory Network of Floral Bud Induction in Longan (Dimocarpus longan Lour.). Front. Plant Sci. 2022, 13, 923183. [Google Scholar] [CrossRef]

| SOYBEAN PART | Treatment (μmol) | SeCys2 | MeSeCys | Se4+ | SeMet | Se6+ | Se_Total |
|---|---|---|---|---|---|---|---|
| root | 0 (L_CK) | ND | ND | 0.03 ± 0.01 c (21.26 ± 4.97) | 0.13 ± 0.02 c (78.74 ± 9.31) | ND | 3.20 ± 0.73 c |
| 20 (L_N20) | 2.25 ± 0.21 b (19.08 ± 1.82) | 2.29 ± 0.29 b (19.48 ± 2.43) | 0.89 ± 0.05 b (7.54 ± 4.33) | 3.90 ± 0.24 b (33.12 ± 2.04) | 2.44 ± 0.06 b (20.77 ± 0.48) | 218.96 ± 22.24 b | |
| 100 (L_N100) | 6.32 ± 0.38 a (22.64 ± 0.13) | 7.22 ± 0.34 a (25.89 ± 1.20) | 1.76 ± 0.07 a (6.30 ± 0.24) | 5.73 ± 0.25 a (20.55 ± 0.90) | 6.87 ± 0.36 a (24.62 ± 1.28) | 518.01 ± 29.82 a | |
| leaf | 0 (R_CK) | ND | ND | 0.04 ± 0.01 c (15.35 ± 3.48) | 0.19 ± 0.02 c (84.65 ± 9.13) | ND | 1.45 ± 0.09 c |
| 20 (R_N20) | 0.19 ± 0.02 b (6.09 ± 0.71) | 0.19 ± 0.05 b (6.01 ± 1.56) | 0.07 ± 0.02 b (2.30 ± 0.75) | 2.18 ± 0.26 b (70.89 ± 0.84) | 0.45 ± 0.00 b (14.71 ± 0.01) | 15.47 ± 2.44 b | |
| 100 (R_N100) | 1.19 ± 0.20 a (17.34 ± 2.90) | 0.95 ± 0.10 a (13.87 ± 1.41) | 0.15 ± 0.03 a (2.14 ± 0.36) | 3.79 ± 0.61 a (55.14 ± 8.89) | 0.79 ± 0.09 a (11.52 ± 1.34) | 42.85 ± 0.39 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Y.; Xiang, X.; Xiao, C.; Zhang, N.; Cheng, H.; Rao, S.; Cheng, S.; Li, L. Illumina RNA and SMRT Sequencing Reveals the Mechanism of Uptake and Transformation of Selenium Nanoparticles in Soybean Seedlings. Plants 2023, 12, 789. https://doi.org/10.3390/plants12040789
Xiong Y, Xiang X, Xiao C, Zhang N, Cheng H, Rao S, Cheng S, Li L. Illumina RNA and SMRT Sequencing Reveals the Mechanism of Uptake and Transformation of Selenium Nanoparticles in Soybean Seedlings. Plants. 2023; 12(4):789. https://doi.org/10.3390/plants12040789
Chicago/Turabian StyleXiong, Yuzhou, Xumin Xiang, Chunmei Xiao, Na Zhang, Hua Cheng, Shen Rao, Shuiyuan Cheng, and Li Li. 2023. "Illumina RNA and SMRT Sequencing Reveals the Mechanism of Uptake and Transformation of Selenium Nanoparticles in Soybean Seedlings" Plants 12, no. 4: 789. https://doi.org/10.3390/plants12040789
APA StyleXiong, Y., Xiang, X., Xiao, C., Zhang, N., Cheng, H., Rao, S., Cheng, S., & Li, L. (2023). Illumina RNA and SMRT Sequencing Reveals the Mechanism of Uptake and Transformation of Selenium Nanoparticles in Soybean Seedlings. Plants, 12(4), 789. https://doi.org/10.3390/plants12040789






