Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter
Abstract
:1. Introduction
2. Results
2.1. Identification of U6 Promoters in Leaf Lettuce
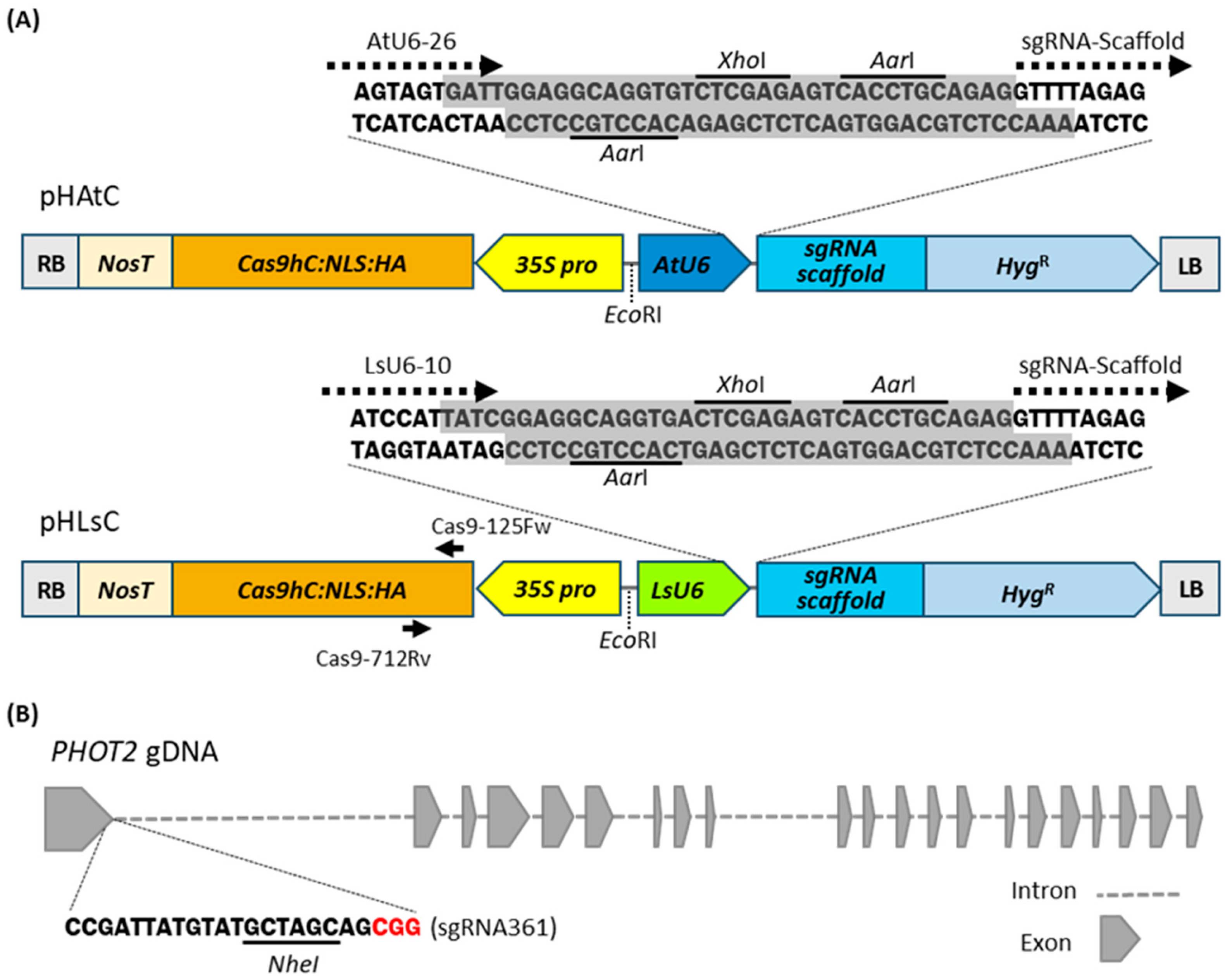
2.2. Plasmid Construction for the CRISPR/Cas9-Mediated Gene Editing System in Lettuce
2.3. The LsU6 Promoter Has Higher Efficiency Than the AtU6 Promoter in CRISPR/Cas9-Mediated Gene Editing in Lettuce
2.4. Lettuce Phot2 Mutants Are Defective in the Chloroplast Avoidance Response
2.5. Selection of Transgene-Free and Genome-Edited Phot2 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning of the LsU6-10 Promoter and Construction of the Binary Vectors
4.3. Agrobacterium-Mediated Lettuce Transformation
4.4. PCR-Based Restriction Enzyme (PCR/RE) Digestion Assay
4.5. Analysis of Chloroplast Photorelocation Movement
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [Green Version]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [Green Version]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef] [Green Version]
- Satheesh, V.; Zhang, H.; Wang, X.; Lei, M. Precise editing of plant genomes–prospects and challenges. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 96, pp. 115–123. [Google Scholar]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Aarabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana Benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Wang, M.-B.; Helliwell, C.A.; Wu, L.-M.; Waterhouse, P.M.; Peacock, W.J.; Dennis, E.S. Hairpin RNAs derived from RNA Polymerase II and Polymerase III promoter-directed transgenes are processed differently in plants. RNA 2008, 14, 903–913. [Google Scholar] [CrossRef] [Green Version]
- Long, L.; Guo, D.-D.; Gao, W.; Yang, W.-W.; Hou, L.-P.; Ma, X.-N.; Miao, Y.-C.; Botella, J.R.; Song, C.-P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, Y.-H.; Sun, X.-J.; Hu, Z.; Jiang, Q.-Y.; Song, G.-H.; Zhang, B.; Zhao, S.-S.; Zhang, H. Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophys. Res. Commun. 2019, 519, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Bernard, G.; Gagneul, D.; Santos, H.A.D.; Etienne, A.; Hilbert, J.-L.; Rambaud, C. Efficient genome editing using CRISPR/Cas9 technology in chicory. Int. J. Mol. Sci. 2019, 20, 1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 52. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Wolabu, T.W.; Park, J.-J.; Chen, M.; Cong, L.; Ge, Y.; Jiang, Q.; Debnath, S.; Li, G.; Wen, J.; Wang, Z. Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 2020, 252, 15. [Google Scholar] [CrossRef]
- Christie, J.M. Phototropin blue-light receptors. Annu Rev Plant Biol 2007, 58, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Kagawa, T.; Sakai, T.; Suetsugu, N.; Oikawa, K.; Ishiguro, S.; Kato, T.; Tabata, S.; Okada, K.; Wada, M. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 2001, 291, 2138–2141. [Google Scholar] [CrossRef]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M.; Okada, K. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef] [Green Version]
- Jarillo, J.A.; Gabrys, H.; Capel, J.; Alonso, J.M.; Ecker, J.R.; Cashmore, A.R. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 2001, 410, 952–954. [Google Scholar] [CrossRef]
- Kim, M.J.; Moon, Y.; Tou, J.C.; Mou, B.; Waterland, N.L. Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.). J. Food Compos. Anal. 2016, 49, 19–34. [Google Scholar] [CrossRef]
- López, A.; Javier, G.-A.; Fenoll, J.; Hellín, P.; Flores, P. Chemical composition and antioxidant capacity of lettuce: Comparative study of regular-sized (Romaine) and baby-sized (Little Gem and Mini Romaine) types. J. Food Compos. Anal. 2014, 33, 39–48. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Wang, S.; Ning, K.; Chen, Z.; Wang, Y.; Yang, J.; Wang, Q. LsAP2 regulates leaf morphology by inhibiting CIN-like TCP transcription factors and repressing LsKAN2 in lettuce. Hortic. Res. 2021, 8, 184. [Google Scholar] [CrossRef]
- Nguyen, C.D.; Li, J.; Mou, B.; Gong, H.; Huo, H. A case study of using an efficient CRISPR/Cas9 system to develop variegated lettuce. Veg. Res. 2021, 1, 4. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.-T.; Ryu, J.; Choi, M.K.; Kweon, J.; Kang, B.-C.; Ahn, H.-M.; Bae, S.; Kim, J.; Kim, J.-S. A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J. Integr. Plant Biol. 2016, 58, 705–712. [Google Scholar] [CrossRef]
- Suetsugu, N.; Kagawa, T.; Wada, M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005, 139, 151–162. [Google Scholar] [CrossRef] [Green Version]
- Hamdan, M.F.; Karlson, C.K.S.; Teoh, E.Y.; Lau, S.-E.; Tan, B.C. Genome editing for sustainable crop improvement and mitigation of biotic and abiotic stresses. Plants 2022, 11, 2625. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Briggs, W.R.; Beck, C.F.; Cashmore, A.R.; Christie, J.M.; Hughes, J.; Jarillo, J.A.; Kagawa, T.; Kanegae, H.; Liscum, E.; Nagatani, A.; et al. The phototropin family of photoreceptors. Plant Cell 2001, 13, 993–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.-W.; Rothfels, C.J.; Melkonian, M.; Villarreal, J.C.; Stevenson, D.W.; Graham, S.W.; Wong, G.K.-S.; Mathews, S.; Pryer, K.M. The origin and evolution of phototropins. Front. Plant Sci. 2015, 6, 637. [Google Scholar] [CrossRef]
- Wada, M.; Kong, S.-G. Analysis of chloroplast movement and relocation in Arabidopsis. In Chloroplast Research in Arabidopsis; Springer: Berlin/Heidelberg, Germany, 2011; pp. 87–102. [Google Scholar]
- Kasahara, M.; Kagawa, T.; Oikawa, K.; Suetsugu, N.; Miyao, M.; Wada, M. Chloroplast avoidance movement reduces photodamage in plants. Nature 2002, 420, 829–832. [Google Scholar] [CrossRef]
- Gotoh, E.; Suetsugu, N.; Yamori, W.; Ishishita, K.; Kiyabu, R.; Fukuda, M.; Higa, T.; Shirouchi, B.; Wada, M. Chloroplast accumulation response enhances leaf photosynthesis and plant biomass production. Plant Physiol. 2018, 178, 1358–1369. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Nelson, R.S.; Sherwood, J.L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 1994, 16, 664–668. [Google Scholar]
- Ismail, H.; Dilshad, E.; Waheed, M.T.; Sajid, M.; Kayani, W.K.; Mirza, B. Transformation of Lactuca sativa L. with rol C gene results in increased antioxidant potential and enhanced analgesic, anti-inflammatory and antidepressant activities in vivo. 3 Biotech 2016, 6, 215. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Riu, Y.-S.; Song, H.-G.; Kim, H.-S.; Kong, S.-G. Guard-cell-specific expression of phototropin2 C-terminal fragment enhances leaf transpiration. Plants 2021, 11, 65. [Google Scholar] [CrossRef]





| Transgenic Line (R0) | NheI Digestion | Mutation Type | Chloroplast Positioning | Indel Pattern |
|---|---|---|---|---|
| phot2-At1 | +/− | Mo | Av | phot2-At1 |
| phot2-At2 | +/− | Mo | Av | phot2-At2 |
| phot2-At3 | − | Bi | Ac | phot2-At3 |
| phot2-At4 | − | Bi | Ac | phot2-At4 |
| phot2-At5 | + | WT | Av | WT |
| phot2-At6 | + | WT | Av | WT |
| phot2-At7 | + | WT | Av | WT |
| phot2-At8 | + | WT | Av | WT |
| phot2-At9 | +/− | Mo | Av | phot2-At1 |
| phot2-At10 | + | WT | Av | WT |
| phot2-At11 | + | WT | Av | WT |
| phot2-At12 | +/− | Mo | Av | phot2-At2 |
| phot2-At13 | + | WT | Av | WT |
| phot2-At14 | +/− | Mo | Av | phot2-At1 |
| phot2-At15 | +/− | Mo | Av | phot2-At1 |
| phot2-At16 | + | WT | Av | WT |
| phot2-At17 | − | Bi | Ac | phot2-At3 |
| phot2-At18 | + | WT | Av | WT |
| phot2-At19 | − | Bi | Ac | phot2-At4 |
| phot2-At20 | + | WT | Av | WT |
| phot2-At21 | + | WT | Av | WT |
| phot2-Ls1 | +/− | Mo | Av | phot2-Ls1 |
| phot2-Ls2 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls3 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls4 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls5 | − | Bi | Ac | phot2-Ls5 |
| phot2-Ls6 | − | Bi | Ac | phot2-Ls6 |
| phot2-Ls7 | − | Bi | Ac | phot2-Ls7 |
| phot2-Ls8 | + | WT | Av | WT |
| phot2-Ls9 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls10 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls11 | + | WT | Av | WT |
| phot2-Ls12 | + | WT | Av | WT |
| phot2-Ls13 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls14 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls15 | − | Bi | Ac | phot2-Ls6 |
| phot2-Ls16 | + | WT | Av | WT |
| phot2-Ls17 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls18 | + | WT | Av | WT |
| phot2-Ls19 | − | Bi | Ac | phot2-Ls5 |
| phot2-Ls20 | + | WT | Av | WT |
| phot2-Ls21 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls22 | − | Bi | Ac | phot2-Ls6 |
| Construct | Number of Transgenic Plants (R0 Generation) | Number of Transgenic Plants with Mutation | Mutation Frequency (%) | Mutation Type | |
|---|---|---|---|---|---|
| Number of Monoallelic (%) | Number of Biallelic (%) | ||||
| pHAtC | 21 | 10 | 48 | 6 (29) | 4 (19) |
| pHLsC | 22 | 16 | 73 | 7 (32) | 9 (41) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riu, Y.-S.; Kim, G.H.; Chung, K.W.; Kong, S.-G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants 2023, 12, 878. https://doi.org/10.3390/plants12040878
Riu Y-S, Kim GH, Chung KW, Kong S-G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants. 2023; 12(4):878. https://doi.org/10.3390/plants12040878
Chicago/Turabian StyleRiu, Young-Sun, Gwang Hoon Kim, Ki Wha Chung, and Sam-Geun Kong. 2023. "Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter" Plants 12, no. 4: 878. https://doi.org/10.3390/plants12040878
APA StyleRiu, Y.-S., Kim, G. H., Chung, K. W., & Kong, S.-G. (2023). Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants, 12(4), 878. https://doi.org/10.3390/plants12040878






