Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees
Abstract
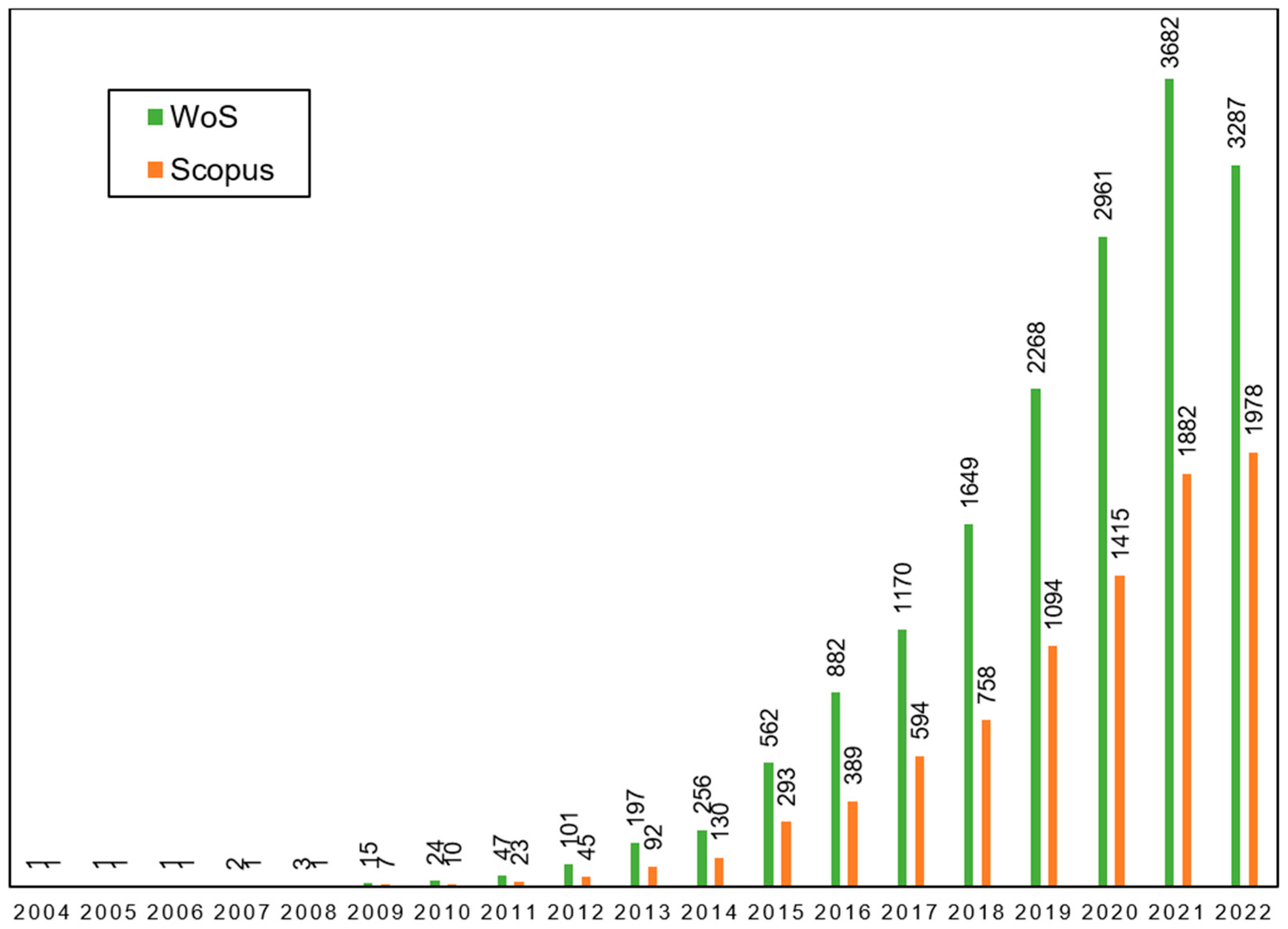
:1. Introduction
2. Olive Tree and Its Vascular Pathogens
3. The Role of Xylem Composition and Its Associated Microorganisms in Plants
4. Methodological Approaches to Study the Xylem Microbiota
4.1. Methods Used to Extract Xylem Microbial Communities
4.2. Assessment of Microbial Communities by Culture-Dependent Approaches
4.3. Assessment of Microbial Communities by Next-Generation Sequencing
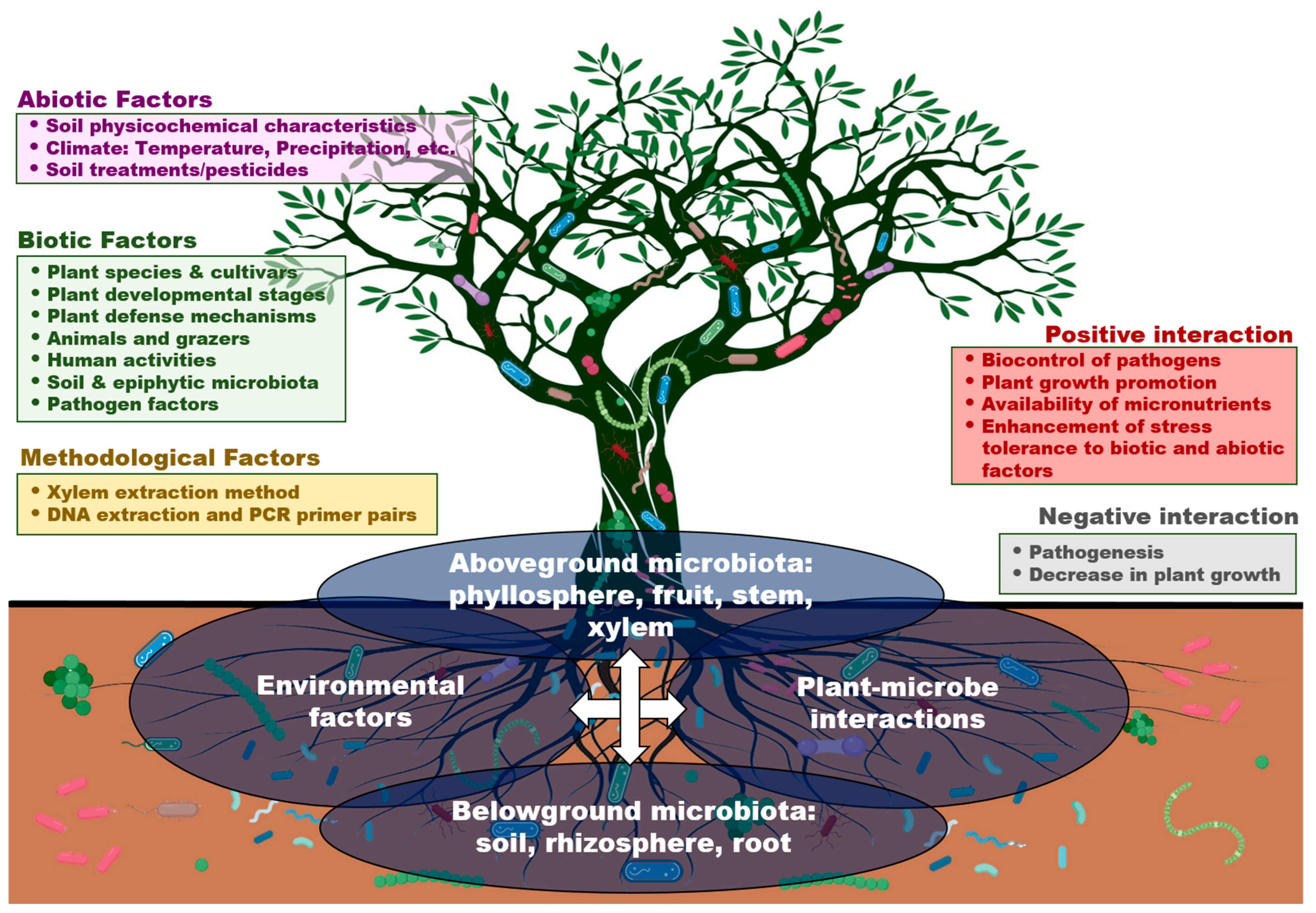
5. Biotic and Abiotic Factors That Influence Xylem Microbiota
5.1. Plant-Associated Factors
5.2. Environmental Factors
5.3. Infection by Vascular Pathogens
| Methods for Extracting the Xylem Microbiome | Reference |
|---|---|
| [57] [57] [58] [57] [39,52,59,60] [40,61] [62] [5,52,60,64] |
| Methods for isolation and identification of microbiome | |
| [52,66,67,68,69,70,71] [85,86,87,88,93,94,95,96] |
| Biotic and abiotic factors | |
| |
| Ecological niche | [114,115,116,117,118,119] |
| Plant age | [39,120,121,122,123,124,125,126] |
| Plant genotype | [39,52,123,144,145] |
| |
| Seasonality | [54,146] |
| Soil type | [147] |
| Soil pH | [148] |
| Drought | [106,149] |
| Agricultural practices | [150] |
| |
| Verticillium dahliae | [15,153] |
| Xylella fastidiosa | [64,154,155,156,157] |
6. Future Perspectives for Xylem Microbiome Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; Gardener, B.B.M.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [Green Version]
- Anguita-Maeso, M.; Trapero-Casas, J.L.; Olivares-García, C.; Ruano-Rosa, D.; Palomo-Ríos, E.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A.; Landa, B.B. Verticillium dahliae inoculation and in vitro propagation modify the xylem microbiome and disease reaction to Verticillium wilt in a wild olive genotype. Front. Plant Sci. 2021, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science (80) 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Song, C.; Zhu, F.; Carrión, V.J.; Cordovez, V. Beyond Plant Microbiome Composition: Exploiting Microbial Functions and Plant Traits via Integrated Approaches. Front. Bioeng. Biotechnol. 2020, 8, 896. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Allen, H.D.; Randall, R.E.; Amable, G.S.; Devereux, B.J. The impact of changing olive cultivation practices on the ground flora of olive groves in the Messara and Psiloritis regions, Crete, Greece. Land Degrad. Dev. 2006, 17, 249–273. [Google Scholar] [CrossRef]
- Gómez, J.A.; Infante-Amate, J.; De Molina, G.M.; Vanwalleghem, T.; Taguas, V.E.; Lorite, I. Olive Cultivation, its Impact on Soil Erosion and its Progression into Yield Impacts in Southern Spain in the Past as a Key to a Future of Increasing Climate Uncertainty. Agriculture 2014, 4, 170–198. [Google Scholar] [CrossRef] [Green Version]
- Tscheulin, T.; Neokosmidis, L.; Petanidou, T.; Settele, J. Influence of landscape context on the abundance and diversity of bees in Mediterranean olive groves. Bull. Entomol. Res. 2011, 101, 557–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Díaz, R.M.; Cirulli, M.; Bubici, G.; del Mar Jiménez-Gasco, M.; Antoniou, P.P.; Tjamos, E.C. Verticillium Wilt, a major threat to olive production: Current status and future prospects for its management. Plant Dis. 2011, 96, 304–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2018, 109, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, R.P.P.; De La Fuente, L.; Koebnik, R.; Lopes, J.R.S.; Parnell, S.; Scherm, H. Addressing the New Global Threat of Xylella fastidiosa. Phytopathology 2019, 109, 172–174. [Google Scholar] [CrossRef] [Green Version]
- Landa, B.B.; Pérez, A.G.; Luaces, P.; Montes-Borrego, M.; Navas-Cortés, J.A.; Sanz, C. Insights into the effect of Verticillium dahliae defoliating-pathotype infection on the content of phenolic and volatile compounds related to the sensory properties of virgin olive oil. Front. Plant Sci. 2019, 10, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Oude Lansink, A. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef] [Green Version]
- Berlanger, I.; Powelson, M.L. Verticillium wilt. Plant Health Instr. 2000, 151, 109–110. [Google Scholar] [CrossRef]
- Báidez, A.G.; Gómez, P.; Del Río, J.A.; Ortuño, A. Dysfunctionality of the xylem in Olea europaea L. Plants associated with the infection process by Verticillium dahliae Kleb. Role of phenolic compounds in plant defense mechanism. J. Agric. Food Chem. 2007, 55, 3373–3377. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J.; Roca, J.M.; Valverde-Corredor, A.; Blanco-López, M.Á. Verticillium wilt of olive in the Guadalquivir Valley (southern Spain): Relations with some agronomical factors and spread of Verticillium dahliae. Phytopathol. Mediterr. 2010, 49, 370–380. [Google Scholar] [CrossRef]
- Bubici, G.; Cirulli, M. Control of Verticillium wilt of olive by resistant rootstocks. Plant Soil 2012, 352, 363–376. [Google Scholar] [CrossRef]
- Jiménez-Fernández, D.; Trapero-Casas, J.L.; Landa, B.B.; Navas-Cortés, J.A.; Bubici, G.; Cirulli, M.; Jiménez-Díaz, R.M. Characterization of resistance against the olive-defoliating Verticillium dahliae pathotype in selected clones of wild olive. Plant Pathol. 2016, 65, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- Montes-Osuna, N.; Mercado-Blanco, J. Verticillium Wilt of Olive and Its Control: What Did We Learn during the Last Decade? Plants 2020, 9, 735. [Google Scholar] [CrossRef] [PubMed]
- Ostos, E.; Garcia-Lopez, M.T.; Porras, R.; Lopez-Escudero, F.J.; Trapero-Casas, A.; Michailides, T.J.; Moral, J. Effect of cultivar resistance and soil management on spatial–temporal development of Verticillium wilt of olive: A long-term study. Front. Plant Sci. 2020, 11, 1595. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D.; et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucci, E.M. Xylella fastidiosa, a new plant pathogen that threatens global farming: Ecology, molecular biology, search for remedies. Biochem. Biophys. Res. Commun. 2018, 502, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Schaad, N.W.; Postnikova, E.; Lacy, G.; Fatmi, M.; Chang, C.-J. Xylella fastidiosa subspecies: X. fastidiosa subsp piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst. Appl. Microbiol. 2004, 27, 763. [Google Scholar] [CrossRef]
- EFSA. Effectiveness of in planta control measures for Xylella fastidiosa. EFSA J. 2019, 17, e05666. [Google Scholar] [CrossRef]
- Di Serio, F.; Bodino, N.; Cavalieri, V.; Demichelis, S.; Di Carolo, M.; Dongiovanni, C.; Fumarola, G.; Gilioli, G.; Guerrieri, E.; Picciotti, U.; et al. Collection of data and information on biology and control of vectors of Xylella fastidiosa. EFSA Support. Publ. 2019, 16, 1628E. [Google Scholar] [CrossRef]
- Morelli, M.; García-Madero, J.M.; Jos, Á.; Saldarelli, P.; Dongiovanni, C.; Kovacova, M.; Saponari, M.; Baños Arjona, A.; Hackl, E.; Webb, S.; et al. Xylella fastidiosa in Olive: A Review of Control Attempts and Current Management. Microorganisms 2021, 9, 1771. [Google Scholar] [CrossRef]
- Landa, B.B.; Saponari, M.; Feitosa-Junior, O.R.; Giampetruzzi, A.; Vieira, F.J.D.; Mor, E.; Robatzek, S. Xylella fastidiosa’s relationships: The bacterium, the host plants and the plant microbiome. New Phytol. 2022, 234, 1598–1605. [Google Scholar] [CrossRef]
- Doty, S.L. Functional importance of the plant microbiome: Implications for agriculture, forestry and bioenergy. Funct. Importance Plant Microbiome. Implic. Agric. For. Bioenergy 2017, 178, 1–111. [Google Scholar] [CrossRef]
- Rabiey, M.; Hailey, L.E.; Roy, S.R.; Grenz, K.; Al-Zadjali, M.A.S.; Barrett, G.A.; Jackson, R.W. Endophytes vs tree pathogens and pests: Can they be used as biological control agents to improve tree health? Eur. J. Plant Pathol. 2019, 155, 711–729. [Google Scholar] [CrossRef] [Green Version]
- De Boer, A.H.; Volkov, V. Logistics of water and salt transport through the plant: Structure and functioning of the xylem. Plant Cell Environ. 2003, 26, 87–101. [Google Scholar] [CrossRef] [Green Version]
- White, P.J. Chapter 3—Long-Distance Transport in the Xylem and Phloem, 3rd ed.; Marschner’s Mineral Nutrition of Higher Plants; Academic Press: San Diego, CA, USA, 2012; pp. 49–70. ISBN 978-0-12-384905-2. [Google Scholar]
- Yadeta, K.; Thomma, B. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurr, U. Xylem sap sampling—New approaches to an old topic. Trends Plant Sci. 1998, 3, 293–298. [Google Scholar] [CrossRef]
- Fernández-García, N.; Hernández, M.; Casado-Vela, J.; Bru, R.; Elortza, F.; Hedden, P.; Olmos, E. Changes to the proteome and targeted metabolites of xylem sap in Brassica oleracea in response to salt stress. Plant Cell Environ. 2011, 34, 821–836. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Natarajan, S.S.; Bennett, J.O.; Sicher, R.C. Protein and metabolite composition of xylem sap from field-grown soybeans (Glycine max). Planta 2011, 233, 921–931. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Haro, C.; Montes-Borrego, M.; De La Fuente, L.; Navas-Cortés, J.A.; Landa, B.B. Metabolomic, ionomic and microbial characterization of olive xylem sap reveals differences according to plant age and genotype. Agronomy 2021, 11, 6. [Google Scholar] [CrossRef]
- Lowe-Power, T.M.; Hendrich, C.G.; von Roepenack-Lahaye, E.; Li, B.; Wu, D.; Mitra, R.; Dalsing, B.L.; Ricca, P.; Naidoo, J.; Cook, D.; et al. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ. Microbiol. 2018, 20, 1330–1349. [Google Scholar] [CrossRef]
- Wallis, C.M.; Chen, J. Grapevine phenolic compounds in xylem sap and tissues are significantly altered during infection by Xylella fastidiosa. Phytopathology 2012, 102, 816–826. [Google Scholar] [CrossRef] [Green Version]
- Rellán-Álvarez, R.; El-Jendoubi, H.; Wohlgemuth, G.; Abadía, A.; Fiehn, O.; Abadía, J.; Alvarez-Fernández, A. Metabolite profile changes in xylem sap and leaf extracts of strategy I plants in response to iron deficiency and resupply. Front. Plant Sci. 2011, 2, 66. [Google Scholar] [CrossRef] [Green Version]
- Gallinger, J.; Gross, J. Unraveling the Host Plant Alternation of Cacopsylla pruni—Adults but Not Nymphs Can Survive on Conifers Due to Phloem/Xylem Composition. Front. Plant Sci. 2018, 9, 484. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Fausto, C.; Mininni, A.N.; Dichio, B.; Lucini, L. Soil management type differentially modulates the metabolomic profile of olive xylem sap. Plant Physiol. Biochem. 2019, 139, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Boutaj, H.; Chakhchar, A.; Meddich, A.; Wahbi, S.; El Alaoui-Talibi, Z.; Douira, A.; Filali-Maltouf, A.; El Modafar, C. Mycorrhizal autochthonous consortium induced defense-related mechanisms of olive trees against Verticillium dahliae. J. Plant Dis. Prot. 2021, 128, 225–237. [Google Scholar] [CrossRef]
- Novelli, S.; Gismondi, A.; Di Marco, G.; Canuti, L.; Nanni, V.; Canini, A. Plant defense factors involved in Olea europaea resistance against Xylella fastidiosa infection. J. Plant Res. 2019, 132, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Fausto, C.; Araniti, F.; Mininni, A.N.; Crecchio, C.; Scagliola, M.; Bleve, G.; Dichio, B.; Sofo, A. Differential olive grove management regulates the levels of primary metabolites in xylem sap. Plant Soil 2021, 460, 281–296. [Google Scholar] [CrossRef]
- McCully, M.E. Niches for bacterial endophytes in crop plants: A plant biologist’s view. Funct. Plant Biol. 2001, 28, 983–990. [Google Scholar] [CrossRef]
- De La Fuente, L.; Merfa, M.V.; Cobine, P.A.; Coleman, J.J. Pathogen Adaptation to the Xylem Environment. Annu. Rev. Phytopathol. 2022, 60, 163–186. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef] [Green Version]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-dependent and culture-independent characterization of the olive xylem microbiota: Effect of sap extraction methods. Front. Plant Sci. 2020, 10, 1708. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.A.; Witzell, J.; Blumenstein, K.; Rozpedowska, E.; Helander, M.; Sieber, T.N.; Gil, L. Resistance to Dutch Elm Disease Reduces Presence of Xylem Endophytic Fungi in Elms (Ulmus spp.). PLoS ONE 2013, 8, e56987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deyett, E.; Rolshausen, P.E. Temporal dynamics of the sap microbiome of grapevine under high Pierce’s Disease pressure. Front. Plant Sci. 2019, 10, 1246. [Google Scholar] [CrossRef] [PubMed]
- Sofo, A.; Mininni, A.N.; Fausto, C.; Scagliola, M.; Crecchio, C.; Xiloyannis, C.; Dichio, B. Evaluation of the possible persistence of potential human pathogenic bacteria in olive orchards irrigated with treated urban wastewater. Sci. Total Environ. 2019, 658, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Vergine, M.; Meyer, J.B.; Cardinale, M.; Sabella, E.; Hartmann, M.; Cherubini, P.; De Bellis, L.; Luvisi, A. The Xylella fastidiosa-resistant olive cultivar “Leccino” has stable endophytic microbiota during the olive quick decline syndrome (OQDS). Pathogens 2020, 9, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexou, M.; Peuke, A. Methods for Xylem Sap Collection. Methods Mol. Biol. 2013, 953, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Passioura, J.B. The Transport of Water from Soil to Shoot in Wheat Seedlings. J. Exp. Bot. 1980, 31, 333–345. [Google Scholar] [CrossRef]
- Haro, C.; Anguita-Maeso, M.; Metsis, M.; Navas-Cortés, J.A.; Landa, B.B. Evaluation of established methods for DNA extraction and primer pairs targeting 16S rRNA gene for bacterial microbiota profiling of olive xylem sap. Front. Plant Sci. 2021, 12, 296. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Haro, C.; Navas-Cortés, J.A.; Landa, B.B. Primer Choice and Xylem-Microbiome-Extraction Method Are Important Determinants in Assessing Xylem Bacterial Community in Olive Trees. Plants 2022, 11, 1320. [Google Scholar] [CrossRef]
- Buhtz, A.; Kolasa, A.; Arlt, K.; Walz, C.; Kehr, J. Xylem sap protein composition is conserved among different plant species. Planta 2004, 219, 610–618. [Google Scholar] [CrossRef]
- Longchar, B.; Phukan, T.; Yadav, S.; Senthil-Kumar, M. An efficient low-cost xylem sap isolation method for bacterial wilt assays in tomato. Appl. Plant Sci. 2020, 8, e11335. [Google Scholar] [CrossRef] [PubMed]
- García-Tejera, O.; López-Bernal, Á.; Orgaz, F.; Testi, L.; Villalobos, F.J. The pitfalls of water potential for irrigation scheduling. Agric. Water Manag. 2021, 243, 106522. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Ares-Yebra, A.; Haro, C.; Román-Écija, M.; Olivares-García, C.; Costa, J.; Marco-Noales, E.; Ferrer, A.; Navas-Cortés, J.A.; Landa, B.B. Xylella fastidiosa infection reshapes microbial composition and network associations in the xylem of almond trees. Front. Microbiol. 2022, 13, 866085. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Anand, G.; Gaur, R.; Yadav, D. Plant-microbiome interactions for sustainable agriculture: A review. Physiol. Mol. Biol. Plants 2021, 27, 165–179. [Google Scholar] [CrossRef]
- Lee, S.A.; Park, J.; Chu, B.; Kim, J.M.; Joa, J.-H.; Sang, M.K.; Song, J.; Weon, H.-Y. Comparative analysis of bacterial diversity in the rhizosphere of tomato by culture-dependent and -independent approaches. J. Microbiol. 2016, 54, 823–831. [Google Scholar] [CrossRef]
- Armanhi, J.S.L.; De Souza, R.S.C.; Damasceno, N.D.B.; de Araújo, L.M.; Imperial, J.; Arruda, P. A Community-Based Culture Collection for Targeting Novel Plant Growth-Promoting Bacteria from the Sugarcane Microbiome. Front. Plant Sci. 2018, 8, 2191. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.I.; Qing, C.; Sze, D.M.-Y.; Roufogalis, B.D.; Neilan, B.A. Culturable Endophytes of Medicinal Plants and the Genetic Basis for Their Bioactivity. Microb. Ecol. 2012, 64, 431–449. [Google Scholar] [CrossRef]
- Shen, S.Y.; Fulthorpe, R. Seasonal variation of bacterial endophytes in urban trees. Front. Microbiol. 2015, 6, 427. [Google Scholar] [CrossRef]
- Yashiro, E.; Spear, R.N.; McManus, P.S. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J. Appl. Microbiol. 2011, 110, 1284–1296. [Google Scholar] [CrossRef]
- Abadi, V.A.J.M.; Sepehri, M.; Rahmani, H.A.; Dolatabad, H.K.; Shamshiripour, M.; Khatabi, B. Diversity and abundance of culturable nitrogen-fixing bacteria in the phyllosphere of maize. J. Appl. Microbiol. 2021, 131, 898–912. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Kiers, E.T. Rewilding plant microbiomes. Science (80) 2022, 378, 599–600. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Liu, H.; Macdonald, C.A.; Singh, B.K. Application of microbial inoculants significantly enhances crop productivity: A meta-analysis of studies from 2010 to 2020. J. Sustain. Agric. Environ. 2022, 1, 216–225. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Ercolani, G.L. Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb. Ecol. 1991, 21, 35–48. [Google Scholar] [CrossRef]
- Aranda, S.; Montes-Borrego, M.; Jiménez-Díaz, R.M.; Landa, B.B. Microbial communities associated with the root system of wild olives (Olea europaea L. subsp. europaea var. sylvestris) are good reservoirs of bacteria with antagonistic potential against Verticillium dahliae. Plant Soil 2011, 343, 329–345. [Google Scholar] [CrossRef] [Green Version]
- Bell, C.R.; Dickie, G.A.; Harvey, W.L.G.; Chan, J.W.Y.F. Endophytic bacteria in grapevine. Can. J. Microbiol. 1995, 41, 46–53. [Google Scholar] [CrossRef]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of Grapevine Flowers, Berries, and Seeds: Identification of Cultivable Bacteria, Comparison with Other Plant Parts, and Visualization of Niches of Colonization. Microb. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef]
- Azevedo, J.; Araújo, W.L.; Lacava, P.T. The diversity of citrus endophytic bacteria and their interactions with Xylella fastidiosa and host plants. Genet. Mol. Biol. 2016, 39, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Durand, A.; Maillard, F.; Alvarez-Lopez, V.; Guinchard, S.; Bertheau, C.; Valot, B.; Blaudez, D.; Chalot, M. Bacterial diversity associated with poplar trees grown on a Hg-contaminated site: Community characterization and isolation of Hg-resistant plant growth-promoting bacteria. Sci. Total Environ. 2018, 622–623, 1165–1177. [Google Scholar] [CrossRef] [Green Version]
- López-Mondéjar, R.; Kostovčík, M.; Lladó, S.; Carro, L.; García-Fraile, P. Exploring the Plant Microbiome Through Multi-Omics Approaches BT—Probiotics in Agroecosystem; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 233–268. ISBN 978-981-10-4059-7. [Google Scholar]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, R.; Boucher, Y.; Dahllöf, I.; Holmström, C.; Doolittle, W.F.; Kjelleberg, S. Use of 16S rRNA and rpoB Genes as Molecular Markers for Microbial Ecology Studies. Appl. Environ. Microbiol. 2007, 73, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, B.E.R.; Sanders, J.G.; Hampton-Marcell, J.; Owens, S.M.; Gilbert, J.A.; Moreau, C.S. DNA extraction protocols cause differences in 16S rRNA amplicon sequencing efficiency but not in community profile composition or structure. Microbiologyopen 2014, 3, 910–921. [Google Scholar] [CrossRef] [Green Version]
- Henderson, G.; Cox, F.; Kittelmann, S.; Miri, V.H.; Zethof, M.; Noel, S.J.; Waghorn, G.C.; Janssen, P.H. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS ONE 2013, 8, e74787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, N.A.; Walker, A.W.; Berry, S.H.; Duncan, S.H.; Farquarson, F.M.; Louis, P.; Thomson, J.M.; Satsangi, J.; Flint, H.J.; Parkhill, J.; et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE 2014, 9, e88982. [Google Scholar] [CrossRef]
- Brooks, J.P.; Edwards, D.J.; Harwich, M.D.; Rivera, M.C.; Fettweis, J.M.; Serrano, M.G.; Reris, R.A.; Sheth, N.U.; Huang, B.; Girerd, P.; et al. The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stulberg, E.; Fravel, D.; Proctor, L.M.; Murray, D.M.; LoTempio, J.; Chrisey, L.; Garland, J.; Goodwin, K.; Graber, J.; Harris, M.C.; et al. An assessment of US microbiome research. Nat. Microbiol. 2016, 1, 15015. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Metagenomics methods for the study of plant-associated microbial communities: A review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef]
- Giangacomo, C.; Mohseni, M.; Kovar, L.; Wallace, J.G. Comparing DNA Extraction and 16S rRNA Gene Amplification Methods for Plant-Associated Bacterial Communities. Phytobiomes J. 2020, 5, 190–201. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comeau, A.M.; Li, W.K.W.; Tremblay, J.-É.; Carmack, E.C.; Lovejoy, C. Arctic Ocean Microbial Community Structure before and after the 2007 Record Sea Ice Minimum. PLoS ONE 2011, 6, e27492. [Google Scholar] [CrossRef] [PubMed]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative Evaluation of Four Bacteria-Specific Primer Pairs for 16S rRNA Gene Surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.; Tringe, S. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Deyett, E.; Roper, M.C.; Ruegger, P.; Yang, J.-I.; Borneman, J.; Rolshausen, P.E. Microbial Landscape of the Grapevine Endosphere in the Context of Pierce’s Disease. Phytobiomes J. 2017, 1, 138–149. [Google Scholar] [CrossRef] [Green Version]
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive orchard microbiome: Characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [Green Version]
- Giampetruzzi, A.; Baptista, P.; Morelli, M.; Cameirão, C.; Lino Neto, T.; Costa, D.; D’Attoma, G.; Abou Kubaa, R.; Altamura, G.; Saponari, M.; et al. Differences in the endophytic microbiome of olive cultivars infected by Xylella fastidiosa across seasons. Pathogens 2020, 9, 723. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Reynolds, H.L.; Packer, A.; Bever, J.D.; Clay, K. Grassroots Ecology: Plant-Microbe-Soil Interactions as Drivers of Plant Community Structure and Dynamics. Ecology 2003, 84, 2281–2291. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [Green Version]
- Bergelson, J.; Mittelstrass, J.; Horton, M.W. Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Eldridge, D.J. Cross-Biome Drivers of Soil Bacterial Alpha Diversity on a Worldwide Scale. Ecosystems 2019, 22, 1220–1231. [Google Scholar] [CrossRef] [Green Version]
- Brunel, C.; Pouteau, R.; Dawson, W.; Pester, M.; Ramirez, K.S.; van Kleunen, M. Towards Unraveling Macroecological Patterns in Rhizosphere Microbiomes. Trends Plant Sci. 2020, 25, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Droby, S.; Wisniewski, M. The fruit microbiome: A new frontier for postharvest biocontrol and postharvest biology. Postharvest Biol. Technol. 2018, 140, 107–112. [Google Scholar] [CrossRef]
- Leveau, J.H.J. A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome. Curr. Opin. Microbiol. 2019, 49, 41–49. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Singh, B. Linking the Phyllosphere Microbiome to Plant Health. Trends Plant Sci. 2020, 25, 841–844. [Google Scholar] [CrossRef]
- Zhang, H.; Serwah Boateng, N.A.; Ngolong Ngea, G.L.; Shi, Y.; Lin, H.; Yang, Q.; Wang, K.; Zhang, X.; Zhao, L.; Droby, S. Unravelling the fruit microbiome: The key for developing effective biological control strategies for postharvest diseases. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4906–4930. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca-García, C.; Coleman-Derr, D.; Garrido, E.; Visel, A.; Tringe, S.G.; Partida-Martínez, L.P. The Cacti Microbiome: Interplay between Habitat-Filtering and Host-Specificity. Front. Microbiol. 2016, 7, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Arias-Giraldo, L.F.; Navas-Cortés, J.A.; de Menezes, A.; Landa, B.B. Bioinformatic pipelines are determinant in the analysis of microbial communities from different ecological niches in cultivated olive trees. In Proceedings of the 4th Annual Conference of the EuroXanth COST Action Integrating Science on Xanthomonadaceae for Integrated Plant Disease Management in Europe, Virtual Conference, 28–30 June 2021; p. 51. Available online: http://hdl.handle.net/10261/268582 (accessed on 10 January 2023).
- Micallef, S.A.; Channer, S.; Shiaris, M.P.; Colon-Carmona, A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 2009, 4, 777–780. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wang, G.; Jin, J.; Liu, J.; Zhang, Q.; Liu, X. Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol. Biochem. 2009, 41, 919–925. [Google Scholar] [CrossRef]
- Marques, J.M.; da Silva, T.F.; Vollu, R.E.; Blank, A.F.; Ding, G.-C.; Seldin, L.; Smalla, K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 2014, 88, 424–435. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [Green Version]
- Caliz, J.; Montes-Borrego, M.; Triadó-Margarit, X.; Metsis, M.; Landa, B.B.; Casamayor, E.O. Influence of edaphic, climatic, and agronomic factors on the composition and abundance of nitrifying microorganisms in the rhizosphere of commercial olive crops. PLoS ONE 2015, 10, e0125787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meaden, S.; Metcalf, C.J.E.; Koskella, B. The effects of host age and spatial location on bacterial community composition in the English Oak tree (Quercus robur). Environ. Microbiol. Rep. 2016, 8, 649–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carper, D.L.; Carrell, A.A.; Kueppers, L.M.; Frank, A.C. Bacterial endophyte communities in Pinus flexilis are structured by host age, tissue type, and environmental factors. Plant Soil 2018, 428, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Marasco, R.; Rolli, E.; Fusi, M.; Michoud, G.; Daffonchio, D. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 2018, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Okayasu, K.; Notaguchi, M. Leaf-associated microbiomes of grafted tomato plants. Sci. Rep. 2019, 9, 1787. [Google Scholar] [CrossRef] [Green Version]
- Montes-Borrego, M.; Jiménez-Díaz, R.M.; Trapero-Casas, J.L.; Navas-Cotés, J.A.; Haro, C.; Rivas, J.C.; De La Fuente, L.; Landa, B.B. Metabolomic characterization of xylem sap of different olive cultivars growing in Spain. In Proceedings of the European Conference on Xylella fastidiosa: Finding Answers to a Global Problem, Palma de Mallorca, Spain, 13–15 November 2017; p. 40. [Google Scholar] [CrossRef]
- Puzanskiy, R.K.; Yemelyanov, V.V.; Shavarda, A.L.; Gavrilenko, T.A.; Shishova, M.F. Age- and Organ-Specific Differences of Potato (Solanum phureja) Plants Metabolome. Russ. J. Plant Physiol. 2018, 65, 813–823. [Google Scholar] [CrossRef]
- Yun, D.-Y.; Kang, Y.-G.; Kim, E.-H.; Kim, M.; Park, N.-H.; Choi, H.-T.; Go, G.H.; Lee, J.H.; Park, J.S.; Hong, Y.-S. Metabolomics approach for understanding geographical dependence of soybean leaf metabolome. Food Res. Int. 2018, 106, 842–852. [Google Scholar] [CrossRef]
- Lawal, U.; Mediani, A.; Maulidiani, M.; Shaari, K.; Ismail, I.S.; Khatib, A.; Abas, F. Metabolite profiling of Ipomoea aquatica at different growth stages in correlation to the antioxidant and α-glucosidase inhibitory activities elucidated by 1H NMR-based metabolomics. Sci. Hortic. 2015, 192, 400–408. [Google Scholar] [CrossRef]
- Zheng, L.; Wang, M.; Ibarra-Estrada, E.; Wu, C.; Wilson, E.; Verpoorte, R.; Klinkhamer, P.; Choi, Y. Investigation of Chemomarkers of Astragali Radix of Different Ages and Geographical Origin by NMR Profiling. Molecules 2015, 20, 3389–3405. [Google Scholar] [CrossRef] [Green Version]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; González-Peña, A.; Peiffer, J.; Koren, O.; Shi, Q.; et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef] [Green Version]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.V.; de Hollander, M.; Garcia, A.A.F.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullington, L.S.; Larkin, B.G. Using direct amplification and next-generation sequencing technology to explore foliar endophyte communities in experimentally inoculated western white pines. Fungal Ecol. 2015, 17, 170–178. [Google Scholar] [CrossRef]
- Yang, R.; Liu, P.; Ye, W. Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Braz. J. Microbiol. 2017, 48, 695–705. [Google Scholar] [CrossRef]
- Montes-Borrego, M.; Navas-Cortés, J.A.; Landa, B.B. Linking microbial functional diversity of olive rhizosphere soil to management systems in commercial orchards in southern Spain. Agric. Ecosyst. Environ. 2013, 181, 169–178. [Google Scholar] [CrossRef] [Green Version]
- Montes-Borrego, M.; Metsis, M.; Landa, B.B. Arbuscular mycorhizal fungi associated with the olive crop across the Andalusian landscape: Factors driving community differentiation. PLoS ONE 2014, 9, e96397. [Google Scholar] [CrossRef] [Green Version]
- Müller, H.; Berg, C.; Landa, B.B.; Auerbach, A.; Moissl-Eichinger, C.; Berg, G. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 2015, 6, 138. [Google Scholar] [CrossRef] [Green Version]
- Morella, N.M.; Weng, F.C.-H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef]
- Leopold, D.R.; Busby, P.E. Host Genotype and Colonist Arrival Order Jointly Govern Plant Microbiome Composition and Function. Curr. Biol. 2020, 30, 3260–3266. [Google Scholar] [CrossRef] [PubMed]
- Anguita-Maeso, M.; Rivas, J.C.; León, G.; Estudillo, C.; Navas-Cortés, J.A.; Landa, B.B. Soil physicochemical properties, seasonality, plant niche and plant genotype affect bacterial and fungal communities in olive orchard soils. In Proceedings of the Global Symposium on Soil Biodiversity, Virtual Conference, 19–22 April 2021; Available online: http://hdl.handle.net/10261/268744 (accessed on 20 December 2022).
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V.; Frederick, M.A. Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. mBio 2017, 8, e00764-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.K.; Millard, P.; Whiteley, A.S.; Murrell, J.C. Unravelling rhizosphere–microbial interactions: Opportunities and limitations. Trends Microbiol. 2004, 12, 386–393. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [Green Version]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Roussos, P.A.; Paplomatas, E.J.; Tjamos, E.C. Phenolic responses of resistant and susceptible olive cultivars induced by defoliating and nondefoliating Verticillium dahliae pathotypes. Plant Dis. 2010, 94, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- D’Attoma, G.; Morelli, M.; Saldarelli, P.; Saponari, M.; Giampetruzzi, A.; Boscia, D.; Savino, V.N.; De La Fuente, L.; Cobine, P.A. Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of Salento, Italy. Pathogens 2019, 8, 272. [Google Scholar] [CrossRef] [Green Version]
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nicolì, F.; Nutricati, E.; Miceli, A.; Negro, C.; de Bellis, L. Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: Profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 2017, 56, 259–273. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Scortichini, M.; Petriccione, M.; Zampella, L.; Mastrobuoni, F.; Cesari, G.; Bertaccini, A.; D’Amico, G.; Contaldo, N.; et al. Xylella fastidiosa and olive quick decline syndrome (CoDiRO) in Salento (southern Italy): A chemometric 1H NMR-based preliminary study on Ogliarola salentina and Cellina di Nardò cultivars. Chem. Biol. Technol. Agric. 2017, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR Metabolite Fingerprinting Analysis Reveals a Disease Biomarker and a Field Treatment Response in Xylella fastidiosa subsp. pauca-Infected Olive Trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Wei, Z.; Wang, X.; Friman, V.-P.; Huang, J.; Wang, X.; Mei, X.; Xu, Y.; Shen, Q.; Jousset, A. Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils 2016, 52, 997–1005. [Google Scholar] [CrossRef] [Green Version]
- Hannula, S.E.; Ma, H.; Pérez-Jaramillo, J.E.; Pineda, A.; Bezemer, T.M. Structure and ecological function of the soil microbiome affecting plant–soil feedbacks in the presence of a soil-borne pathogen. Environ. Microbiol. 2020, 22, 660–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabella, E.; Aprile, A.; Genga, A.; Siciliano, T.; Nutricati, E.; Nicolì, F.; Vergine, M.; Negro, C.; De Bellis, L.; Luvisi, A. Xylem cavitation susceptibility and refilling mechanisms in olive trees infected by Xylella fastidiosa. Sci. Rep. 2019, 9, 9602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Heijden, M.G.A.; Hartmann, M. Networking in the Plant Microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.K.; Kim, H.; Lee, Y.-H. Cross-kingdom co-occurrence networks in the plant microbiome: Importance and ecological interpretations. Front. Microbiol. 2022, 13, 953300. [Google Scholar] [CrossRef]
- Enespa; Chandra, P. Tool and techniques study to plant microbiome current understanding and future needs: An overview. Commun. Integr. Biol. 2022, 15, 209–225. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Lee, K.K.; Jeon, J.; Harris, W.A.; Lee, Y.-H. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 2020, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Nowrotek, M.; Jałowiecki, Ł.; Harnisz, M.; Płaza, G.A. Culturomics and metagenomics: In understanding of environmental resistome. Front. Environ. Sci. Eng. 2019, 13, 40. [Google Scholar] [CrossRef] [Green Version]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Mapelli, F.; Mengoni, A.; Riva, V.; Borin, S. Bacterial culturing is crucial to boost sustainable agriculture. Trends Microbiol. 2022, 31, 1–4. [Google Scholar] [CrossRef]
- Stanley, C.E.; Grossmann, G.; i Solvas, X.C.; deMello, A.J. Soil-on-a-Chip: Microfluidic platforms for environmental organismal studies. Lab Chip 2016, 16, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Abt, B.; Sikorski, J. Present and Future of Culturing Bacteria. Annu. Rev. Microbiol. 2017, 71, 711–730. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, M.S.; Hamza, M.A.; Youssef, H.H.; Patz, S.; Becker, M.; ElSawey, H.; Nemr, R.; Daanaa, H.-S.A.; Mourad, E.F.; Morsi, A.T.; et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media—A review. J. Adv. Res. 2019, 19, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Anguita-Maeso, M.; Olivares-García, C.; Rivas, J.C.; Navas-Cortés, J.A.; Landa, B.B. Broth media cultivation of xylem microbiome from cultivated olive trees. In Proceedings of the III European Conference on Xylella fastidiosa, Virtual Conference, 26–30 April 2021. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; León-Ropero, G.; Trapero-Casas, J.L.; Navas-Cortés, J.A.; Landa, B.B. Plant endotherapy treatments enable the modification of xylem microbiome composition in olive trees. In Proceedings of the 9th IOBC-WPRS Meeting on Integrated Protection of Olive Crops, 26–29 October 2021; p. 59. Available online: http://hdl.handle.net/10261/268480 (accessed on 12 January 2023).
- Berger, G.; Czarnocka, K.; Cochard, B.; Oszako, T.; Lefort, F. Biocontrol endotherapy with Trichoderma spp. and Bacillus amyloliquefaciens against Phytophthora spp.: A comparative study with phosphite treatment on Quercus robur and Fagus sylvatica. J. Agric. Sci. Technol. 2015, 5, 428–439. [Google Scholar] [CrossRef] [Green Version]
- Postma, J.; Goossen-van de Geijn, H. Twenty-four years of Dutch Trig® application to control Dutch elm disease. BioControl 2016, 61, 305–312. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef]
- Faino, L.; Scala, V.; Albanese, A.; Modesti, V.; Grottoli, A.; Pucci, N.; Doddi, A.; L’Aurora, A.; Tatulli, G.; Reverberi, M.; et al. Nanopore sequencing for the detection and identification of Xylella fastidiosa subspecies and sequence types from naturally infected plant material. Plant Pathol. 2021, 70, 1860–1870. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anguita-Maeso, M.; Navas-Cortés, J.A.; Landa, B.B. Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees. Plants 2023, 12, 912. https://doi.org/10.3390/plants12040912
Anguita-Maeso M, Navas-Cortés JA, Landa BB. Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees. Plants. 2023; 12(4):912. https://doi.org/10.3390/plants12040912
Chicago/Turabian StyleAnguita-Maeso, Manuel, Juan A. Navas-Cortés, and Blanca B. Landa. 2023. "Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees" Plants 12, no. 4: 912. https://doi.org/10.3390/plants12040912
APA StyleAnguita-Maeso, M., Navas-Cortés, J. A., & Landa, B. B. (2023). Insights into the Methodological, Biotic and Abiotic Factors Influencing the Characterization of Xylem-Inhabiting Microbial Communities of Olive Trees. Plants, 12(4), 912. https://doi.org/10.3390/plants12040912






