Pre- and Post-Zygotic Barriers Contribute to Reproductive Isolation and Correlate with Genetic Distance in Cucumis
Abstract
:1. Introduction
2. Results
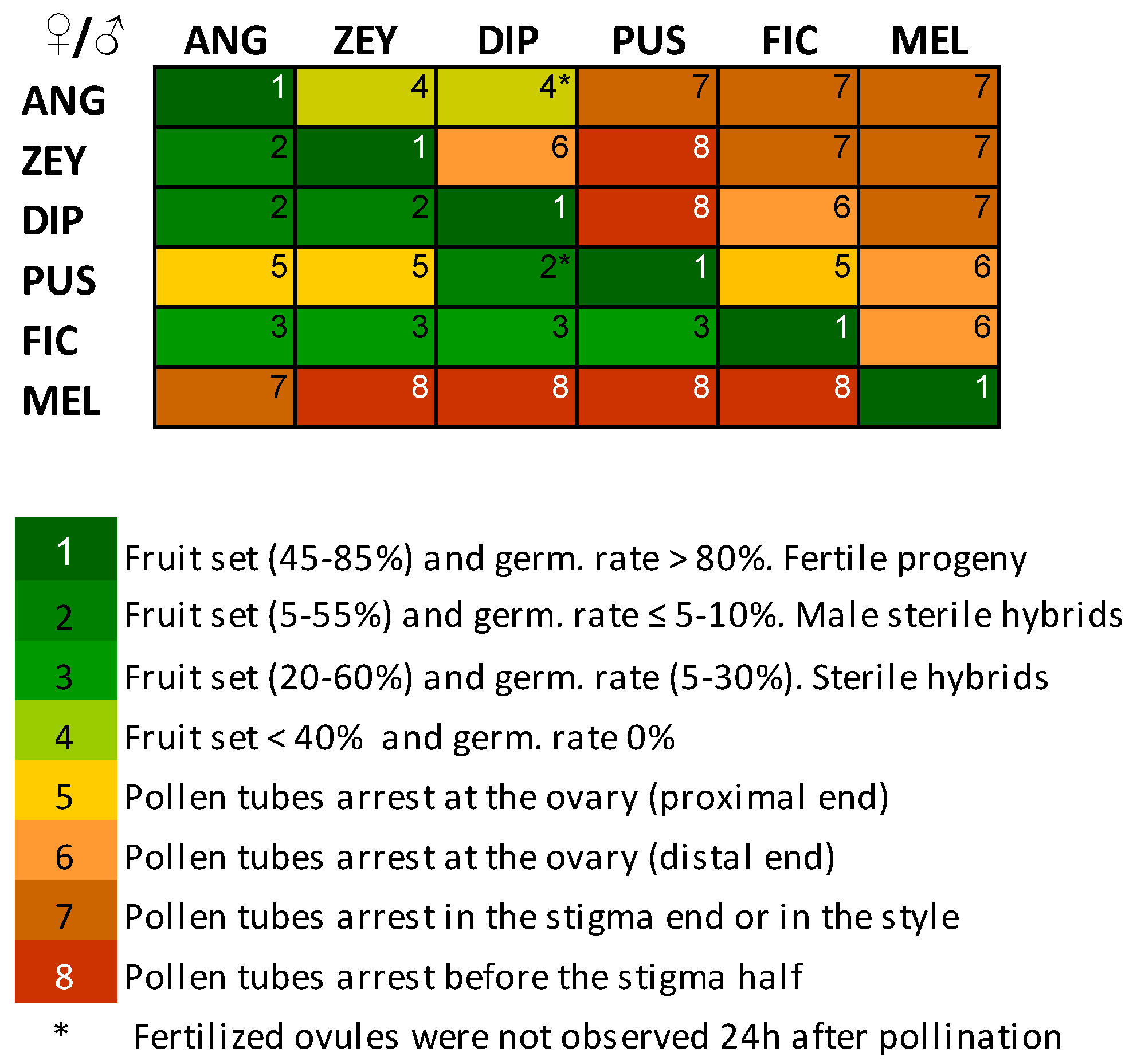
2.1. Patterns of Crossability in Cucumis
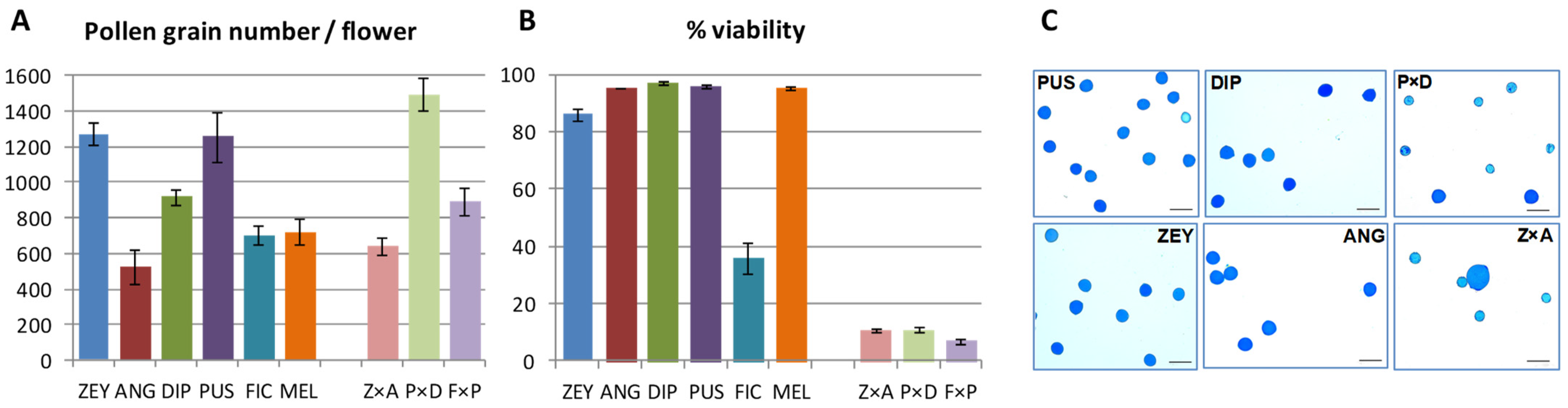
2.2. Pre-Mating Factors Do Not Correlate with the Average Crossability Index
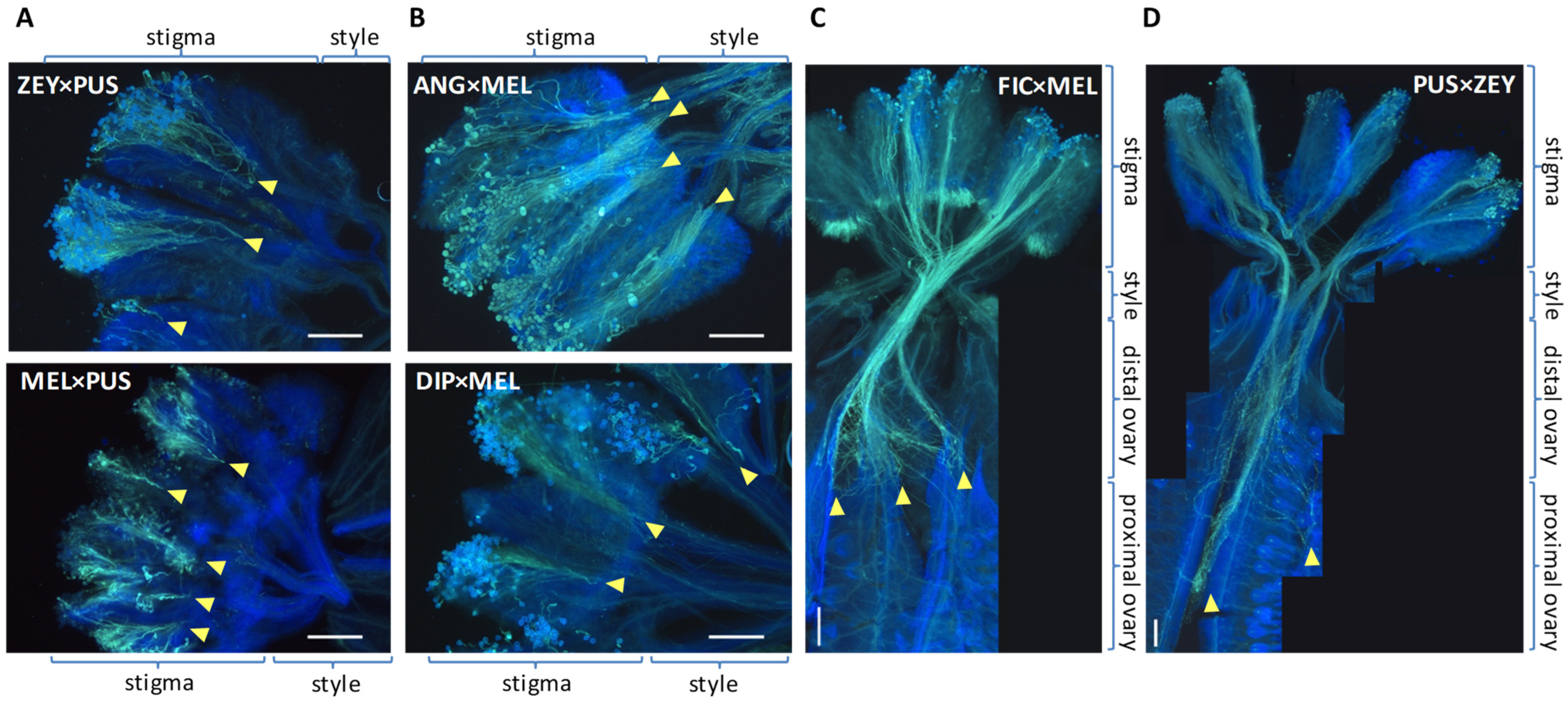
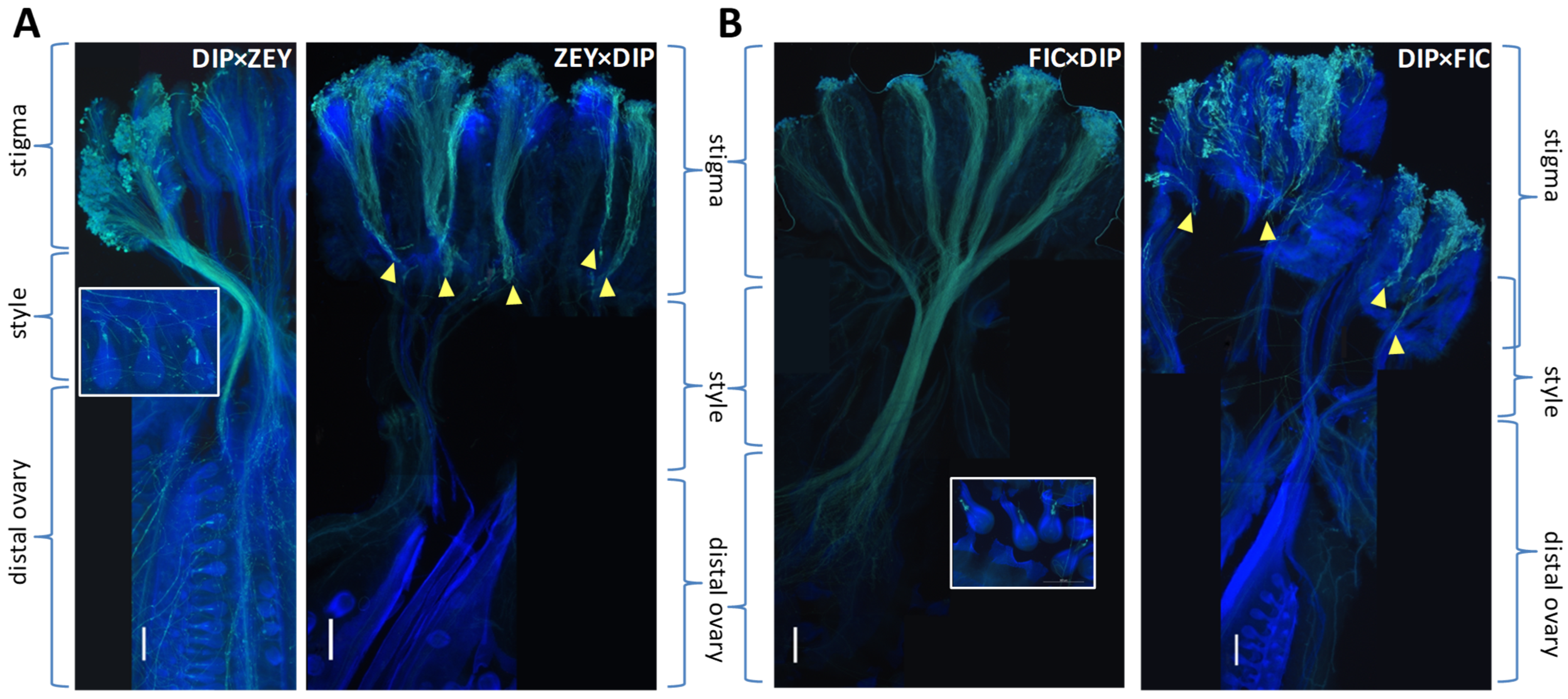
2.3. Pre-Zygotic IRBs in Cucumis Act Predominantly at the Stigma and at the Ovary
2.4. Pollen Tube Arrest and Unilateral Cross-Incongruity/Incompatibility
2.5. Post-Zygotic IRBs: Fruit Set, Seed Set, and Seed Viability
2.6. Post-Zygotic IRBs: Fertility in Interspecific Hybrids
2.7. Pre- and Post-Zygotic IRBs Contribute to Reproductive Isolation (RI) in Cucumis
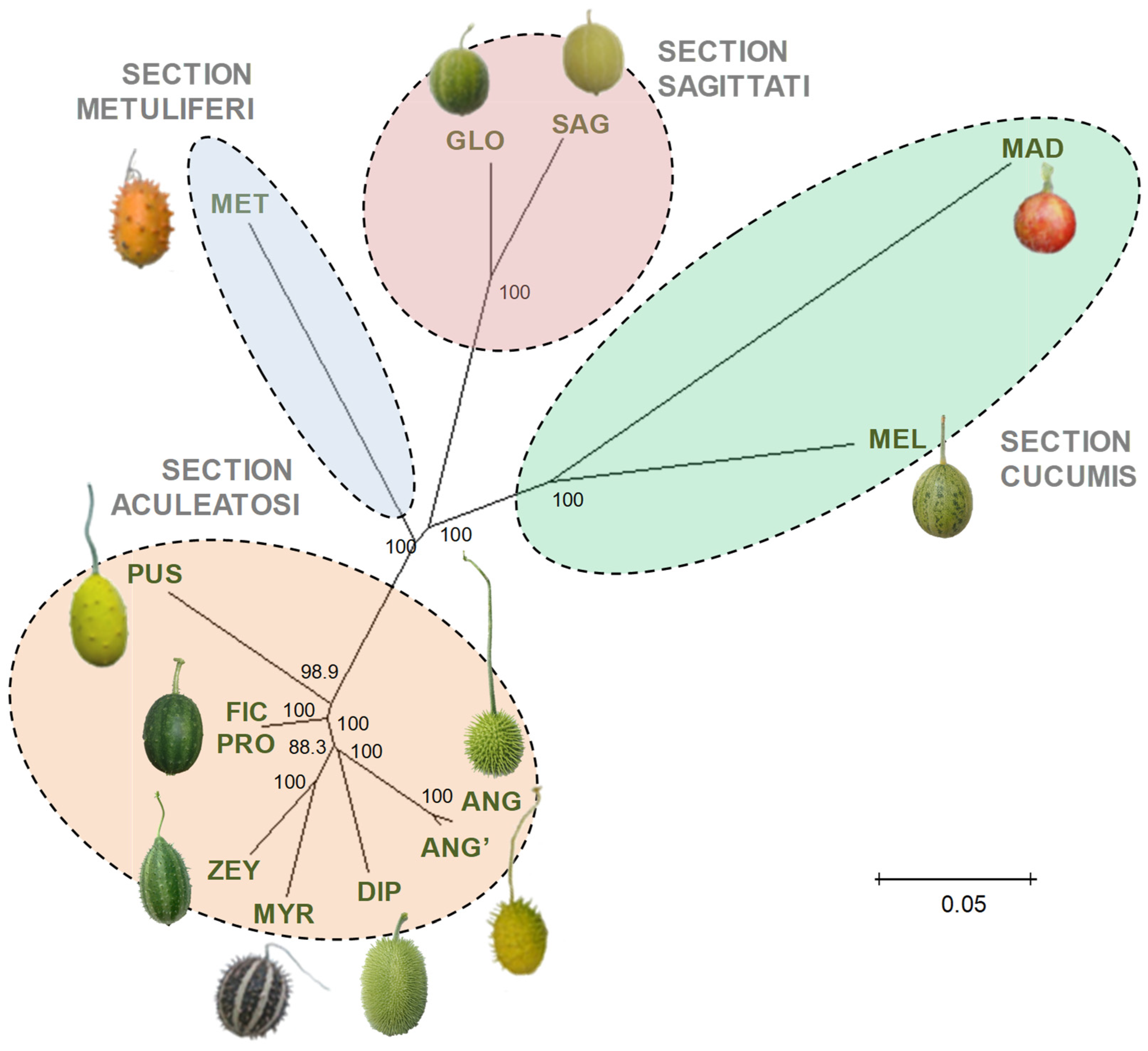
2.8. Genetic Diversity and Phylogenetic Clustering Analysis
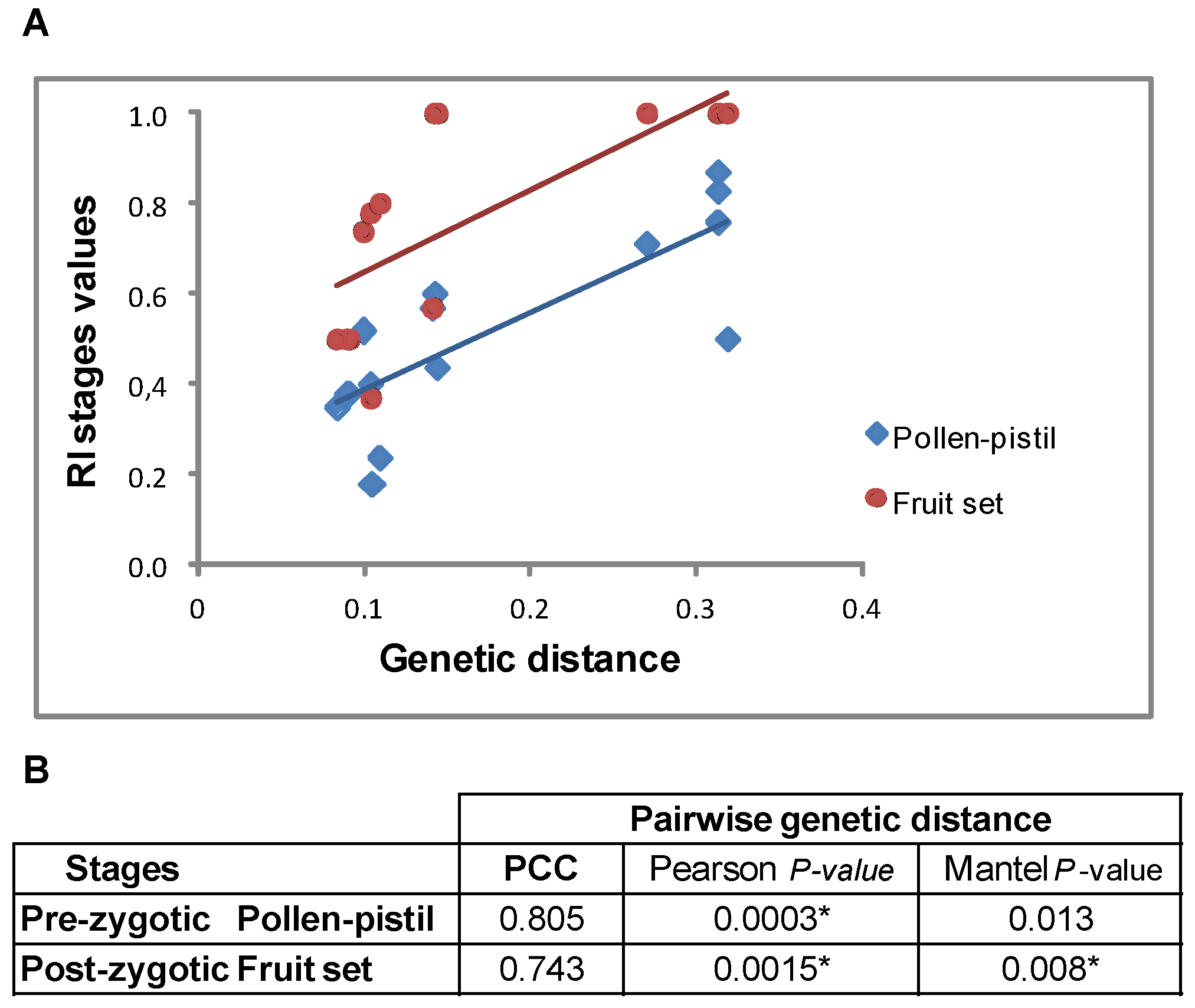
2.9. Correlation between Pre- and Post-Zygotic Isolation and Genetic Distance
3. Discussion
3.1. Pre-Zygotic IRBs in Cucumis
3.2. Post-Zygotic IRBs in Cucumis
3.3. Reproductive Isolation and Genetic Distance
4. Materials and Methods
4.1. Plant Material
4.2. Plant Cultivation and Handling
4.3. Reproductive Isolation Measures
4.4. Fluorescence Microscopy
4.5. DNA Extraction, Ploidy Detection, and SSR Analyses
4.6. Genotyping-by-Sequencing
4.7. Phylogenetic Analysis
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Sebastian, P.; Schaefer, H.; Telford, I.R.H.; Renner, S.S. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. USA 2010, 107, 14269–14273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkbride, J.H. Biosystematic Monograph of the Genus Cucumis (Cucurbitaceae); Parkway Publishers: Boone, NC, USA, 1993. [Google Scholar]
- Kho, Y.O.; Den Nijs, A.P.M.; Franken, J. Interspecific hybridization in Cucumis L. II. The crossability of species, an investigation of in vivo pollen tube growth and seed set. Euphytica 1980, 29, 661–672. [Google Scholar] [CrossRef]
- Barrett, S.C.H. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002, 3, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Igic, B.; Lande, R.; Kohn, J. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 2008, 169, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.S. Pathways for making unisexual flowers and unisexual plants: Moving beyond the “two mutations linked on one chromosome” model. Am. J. Bot. 2016, 103, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Bertin, R.I. Incidence of monoecy and dichogamy in relation to self-fertilization in angiosperms. Am. J. Bot. 1993, 80, 557–560. [Google Scholar] [CrossRef]
- Chen, J.F.; Zhou, X.H. Cucumis. In Wild Crop Relatives: Genomic and Breeding Resources: Vegetables; Springer: Berlin/Heidelberg, Germany, 2011; pp. 67–90. [Google Scholar]
- den Nijs, A.P.M.; Visser, D.L. Relationships between African species of the genus Cucumis L. estimated by the production, vigour and fertility of F1 hybrids. Euphytica 1985, 34, 279–290. [Google Scholar] [CrossRef]
- Raamsdonk, L.W.D.; Den Nijs, A.P.M.; Jongerius, M.C. Meiotic analyses of Cucumis hybrids and an evolutionary evaluation of the genus Cucumis (Cucurbitaceae). Plant Syst. Evol. 1989, 163, 133–146. [Google Scholar] [CrossRef]
- Kishi, Y.; Fujishita, N. Studies on interspecific hybridization in the genus Cucumis. II Pollen tube growth, fertilization and embryogenesis of post-fertilization stage in incompatible crossings. J. Jap. Soc. Hort. Sci. 1970, 39, 51–57. [Google Scholar]
- Singh, A.K.; Yadava, K.S. An analysis of interspecific hybrids and phylogenetic implications in Cucumis (Cucurbitaceae). Plant Syst. Evol. 1984, 147, 237–252. [Google Scholar] [CrossRef] [Green Version]
- Deakin, J.R.; Bohn, G.W.; Whitaker, T.W. Interspecific hybridization in Cucumis. Econ. Bot. 1971, 25, 195–211. [Google Scholar] [CrossRef]
- Norton, J.D.; Granberry, D.M. Characteristics of progeny from an interspecific cross of Cucumis melo with C. metuliferus. J. Am. Soc. Hort. Sci. 1980, 105, 174–180. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Miyagi, M.; Watanabe, N.; Kuboyama, T. Temperature-dependent enhancement of pollen tube growth observed in interspecific crosses between wild Cucumis spp. and melon (C. melo L.). Sci. Hort. 2012, 138, 144–150. [Google Scholar] [CrossRef]
- Chen, J.F.; Staub, J.E.; Tashiro, Y.; Isshiki, S.; Miyazaki, S. Successful interspecific hybridization between Cucumis sativus L. and C. hystrix Chakr. Euphytica 1997, 96, 413–419. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhu, W.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef] [PubMed]
- Bedinger, P.A.; Chetelat, R.T.; McClure, B.; Moyle, L.C.; Rose, J.K.C.; Stack, S.M.; van der Knaap, E.; Baek, Y.S.; Lopez-Casado, G.; Covey, P.A.; et al. Interspecific reproductive barriers in the tomato clade: Opportunities to decipher mechanisms of reproductive isolation. Sex. Plant Reprod. 2010, 24, 171–187. [Google Scholar] [CrossRef] [PubMed]
- García-Más, J.; Monforte, A.J.; Arús, P. Phylogenetic relationships among Cucumis species based on the ribosomal internal transcribed spacer sequence and microsatellite markers. Plant Syst. Evol. 2004, 248, 191–203. [Google Scholar] [CrossRef]
- Renner, S.S.; Schaefer, H.; Kocyan, A. Phylogenetics of Cucumis (Cucurbitaceae): Cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Evol. Biol. 2007, 7, 58. [Google Scholar] [CrossRef] [Green Version]
- Ghebretinsae, A.G.; Thulin, M.; Barber, J.C. Relationships of cucumbers and melons unraveled: Molecular phylogenetics of Cucumis and related genera (Benincaseae, Cucurbitaceae). Am. J. Bot. 2007, 94, 1256–1266. [Google Scholar] [CrossRef]
- Endl, J.; Achigan-Dako, E.G.; Pandey, A.K.; Monforte, A.J.; Pico, B.; Schaefer, H. Repeated domestication of melon (Cucumis melo) in Africa and Asia and a new close relative from India. Am. J. Bot. 2018, 105, 1662–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsey, J.; Bradshaw, H.D.; Schemske, D.W. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phyrmaceae). Evolution 2003, 57, 1520–1534. [Google Scholar] [PubMed]
- African Plant Database (version 4.0.0). Conservatoire et Jardin botaniques de la Ville de Genève and South African National Biodiversity Institute, Pretoria. Available online: http://africanplantdatabase.ch (accessed on 18 January 2023).
- Williams, E.G.; Rouse, J.L. Relationships of pollen size, pistil length and pollen tube growth rate in Rhododendron and their influence on hybridization. Sex. Plant Reprod. 1990, 3, 7–17. [Google Scholar] [CrossRef]
- Aguilar, R.; Bernardello, G.; Galetto, L. Pollen–pistil relationships and pollen size-number trade-off in species of the tribe Lycieae (Solanaceae). J. Plant Res. 2002, 115, 335–340. [Google Scholar] [CrossRef]
- Kitashiba, H.; Nasrallah, J.B. Self-incompatibility in Brassicaceae crops: Lessons for interspecific incompatibility. Breed Sci. 2014, 64, 23–37. [Google Scholar] [CrossRef] [Green Version]
- Kandasamy, M.K.; Nasrallah, J.B.; Nasrallah, M.E. Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 1994, 120, 3405–3418. [Google Scholar] [CrossRef]
- Covey, P.A.; Kondo, K.; Welch, L.; Frank, E.; Sianta, S.; Kumar, A.; Nuñez, R.; Lopez-Casado, G.; van der Knaap, E.; Rose, J.K.C.; et al. Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade. Plant J. 2010, 64, 367–378. [Google Scholar] [CrossRef]
- Murfett, J.; Strabala, T.J.; Zurek, D.M.; Mou, B.; Beecher, B.; McClure, B.A. S-RNase and interspecific pollen rejection in the Genus Nicotiana: Multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and selfcompatible species. Plant Cell 1996, 8, 943–958. [Google Scholar] [CrossRef]
- De Nettancourt, D. Incompatibility and Incongruity in Wild and Cultivated Species, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2001. [Google Scholar]
- Lewis, D.; Crowe, L. Unilateral interspecific incompatibility in flowering plants. Heredity 1958, 12, 233–256. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Li, W.; Liu, Y.; Tan, M.; Ganal, M.; Chetelat, R.T. A farnesyl pyrophosphate synthase gene expressed in pollen functions in S-RNase independent unilateral incompatibility. Plant J. 2018, 93, 417–430. [Google Scholar] [CrossRef]
- Hogenboom, N.G. A model for incongruity in intimate partner relationships. Euphytica 1973, 22, 219–233. [Google Scholar] [CrossRef]
- Dresselhaus, T.; Lausser, A.; Marton, M.L. Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. 2011, 108, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, B.; Chen, Z.; Zhang, D.; Zhang, H.; Wang, H.; Zhang, Y.; Cai, D.; Liu, J.; Xiao, S.; et al. A pectin methylesterase gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nat. Comm. 2018, 9, 3678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kermicle, J.L.; Evans, M.S.S. Pollen-pistil barriers to crossing in maize and teosinte result from incongruity rather than active rejection. Sex Plant Reprod. 2005, 18, 187–194. [Google Scholar] [CrossRef]
- Moyle, L.C.; Graham, E.B. Genetics of Hybrid Incompatibility Between Lycopersicon esculentum and L. hirsutum. Genetics 2007, 169, 355–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, B.A.; Cruz-García, F.; Beecher, B.; Sulaman, W. Factors Affecting Inter- and Intra-specific Pollen Rejection in Nicotiana. Ann. Bot. 2000, 85, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Jewell, J.; Papineau, A.D.; Freyre, R.; Moyle, L. Patterns of reproductive isolation in Nolana (Chilean bellflower). Evolution 2012, 66, 2628–2636. [Google Scholar] [CrossRef]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Baack, E.; Melo, M.C.; Rieseberg, L.H.; Ortiz-Barrientos, D. The origins of reproductive isolation in plants. New Phytol. 2015, 207, 968–984. [Google Scholar] [CrossRef]
- Barringer, B.C. Polyploidy and Self-Fertilization in Flowering Plants. Am. J. Bot. 2007, 94, 1527–1533. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizintin, L.; Bohanec, B. In vitro manipulation of cucumber (Cucumis sativus L.) pollen and microspores: Isolation procedures, viability tests, germination, maturation. Acta Biol. Crac. Ser. Bot. 2004, 46, 177–183. [Google Scholar]
- World Flora Online. Available online: http://www.worldfloraonline.org (accessed on 22 June 2022).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Danin-Poleg, Y.; Rets, N. Development and characterization of microsatellite markers in Cucumis. Theor. Appl. Genet. 2001, 102, 61–72. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labelling of PCR fragments. Nat. Biotech. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Knaus, B.J.; Grünwald, N.J. VCFR: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Res. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 22 June 2022).
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 22 June 2022).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. R Package Version 1.0.5. 2021. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 22 June 2022).
- Metsalu, T.; Vilo, J. Clustvis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef] [PubMed]




| Mean Contribution a | ||||
|---|---|---|---|---|
| Sequencial Stages | N | Absolute | Relative | |
| Pre-zygotic | Pollen-pistil | 15 | 0.515 | 0.519 |
| Post-zygotic | Fruit set | 8 | 0.289 | 0.291 |
| Fruit weigth | 8 | 0.110 | 0.111 | |
| Seed viability | 8 | 0.078 | 0.079 | |
| Total Post-zygotic | 8 | 0.477 | 0.481 | |
| Total RI | 0.992 | 1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferriol, M.; Simó, U.; Mansanet, C.J.; Torres, A.; Picó, B.; Monforte, A.J.; Romero, C. Pre- and Post-Zygotic Barriers Contribute to Reproductive Isolation and Correlate with Genetic Distance in Cucumis. Plants 2023, 12, 926. https://doi.org/10.3390/plants12040926
Ferriol M, Simó U, Mansanet CJ, Torres A, Picó B, Monforte AJ, Romero C. Pre- and Post-Zygotic Barriers Contribute to Reproductive Isolation and Correlate with Genetic Distance in Cucumis. Plants. 2023; 12(4):926. https://doi.org/10.3390/plants12040926
Chicago/Turabian StyleFerriol, María, Unzué Simó, Carme J. Mansanet, Alejandro Torres, Belén Picó, Antonio J. Monforte, and Carlos Romero. 2023. "Pre- and Post-Zygotic Barriers Contribute to Reproductive Isolation and Correlate with Genetic Distance in Cucumis" Plants 12, no. 4: 926. https://doi.org/10.3390/plants12040926






