Disentangling Relationships among the Alpine Species of Luzula Sect. Luzula (Juncaceae) in the Eastern Alps
Abstract
1. Introduction
2. Results
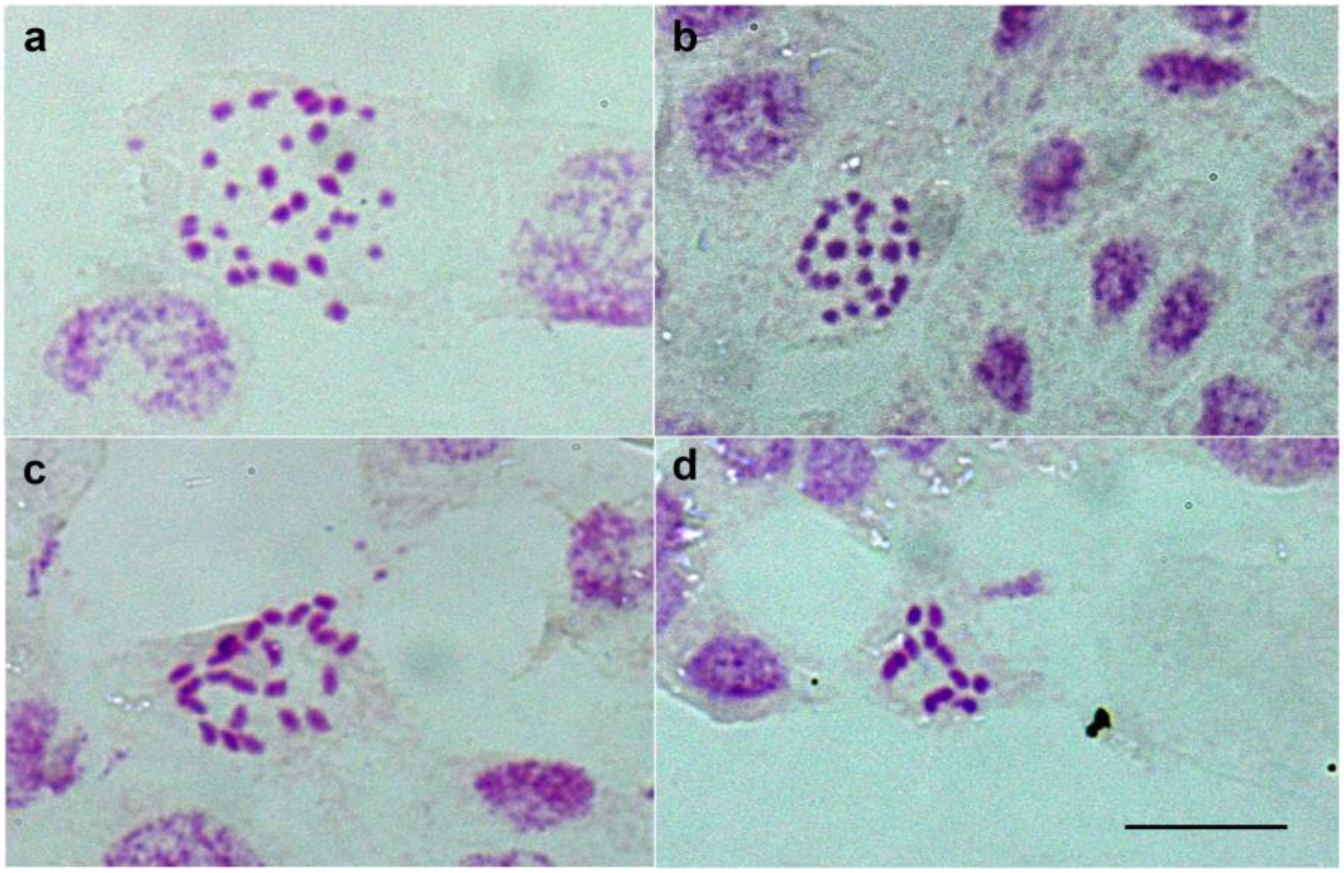
2.1. Karyotype Variation
2.2. Relative Genome Size and Ploidy Levels
2.3. AFLP Fingerprinting and Analyses of AFLP Data
3. Discussion
3.1. Monophyletic Origin of Agmatoploid L. exspectata and L. sudetica
3.2. Recurrent Formation of Tetraploids and Agmatoploids in the L. alpina/L. multiflora Complex
4. Materials and Methods
4.1. Plant Material and Taxon Identification
4.2. Karyotype Determination
4.3. Relative Genome Size Estimation
4.4. AFLP Fingerprinting and Analyses of AFLP Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soltis, D.E.; Albert, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 96, 336–348. [Google Scholar] [CrossRef]
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 2009, 106, 13875–13879. [Google Scholar] [CrossRef]
- Husband, B.C.; Baldwin, S.J.; Suda, J. The incidence of polyploidy in natural plant populations: Major patterns and evolutionary processes. In Plant Genome Diversity; Leitch, I.J., Greilhuber, J., Doležel, J., Wendel, J.F., Eds.; Springer: New York, NY, USA, 2013; Volume 2, pp. 255–276. [Google Scholar] [CrossRef]
- Madlung, A. Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity 2013, 110, 99–104. [Google Scholar] [CrossRef]
- Leitch, I.J.; Bennett, M.D. Genome downsizing in polyploid plants. Biol. J. Linn. Soc. 2004, 82, 651–663. [Google Scholar] [CrossRef]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Sonnleitner, M.; Flatscher, R.; Escobar García, P.; Rauchová, J.; Suda, J.; Schönswetter, P. Distribution and habitat segregation on different spatial scales among diploid, tetraploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps. Ann. Bot. 2010, 106, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Balao, F.; Herrera, J.; Talavera, S. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: A multivariate morphological approach. New Phytol. 2011, 192, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Weiss-Schneeweiss, H.; Emadzade, K.; Jang, T.-S.; Schneeweiss, G.M. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet. Genome Res. 2013, 140, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Soltis, D.E.; Visger, C.J.; Blaine Marchant, D.; Soltis, P.S. Polyploidy: Pitfalls and paths to a paradigm. Am. J. Bot. 2016, 103, 1146–1166. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Rees, H. Nuclear DNA and evolution of allotetraploid Brassicae. Heredity 1974, 33, 61–68. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Kovarik, A.; Kelly, L.J.; Macas, J.; Novak, P.; Chase, M.W.; Nichols, R.A.; Pancholi, M.R.; Grandbastien, M.A.; Leitch, A.R. Diploidization and genome size change in allopolyploids is associated with differential dynamics of low- and highcopy sequences. Plant J. 2013, 74, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Mello-Sampayo, T. Differential polyteny and karyotype evolution in “Luzula”, a critical interpretation of morphological and cytophotometric data. Genética Ibérica 1961, 13, 1–22. [Google Scholar]
- Bačič, T.; Jogan, N.; Dolenc Koce, J. Luzula sect. Luzula in the south-eastern Alps-karyology and genome size. Taxon 2007, 56, 129–136. [Google Scholar] [CrossRef]
- Malheiros, N.; Gardé, A. Fragmentation as a possible evolutionary process in the genus Luzula DC. Genética Ibérica 1950, 2, 257–262. [Google Scholar]
- Löve, A.; Löve, D.; Raymond, M. Cytotaxonomy of Carex section Capillares. Can. J. Bot. 1957, 35, 715–776. [Google Scholar] [CrossRef]
- Bureš, P.; Zedek, F.; Marková, M. Holocentric chromosomes. In Plant Genome Diversity. Physical Structure of Plant Genomes; Wendel, J., Greilhuber, J., Doležel, J., Leitch, I.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2. [Google Scholar] [CrossRef]
- Nordenskiöld, H. Cytotaxonomical studies in the genus Luzula I. Somatic chromosomes and chromosome numbers. Hereditas 1951, 37, 325–355. [Google Scholar] [CrossRef]
- Haizel, T.; Lim, Y.K.; Leitch, A.R.; Moore, G. Molecular analysis of holocentric centromeres of Luzula species. Cytogenet. Genome Res. 2005, 109, 134–143. [Google Scholar] [CrossRef]
- Nagaki, K.; Kashihara, K.; Murata, M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula Nivea. Plant Cell 2005, 17, 1886–1893. [Google Scholar] [CrossRef]
- Bozek, M.; Leitch, A.R.; Leitch, I.J.; Záveská Drábková, L.; Kuta, E. Chromosome and genome size variation in Luzula (Juncaceae), a genus with holocentric chromosomes. Bot. J. Linn. Soc. 2012, 170, 529–541. [Google Scholar] [CrossRef]
- Guerra, M. Agmatoploidy and symploidy: A critical review. Genet. Mol. Biol. 2016, 39, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Luceño, M.; Guerra, M. Numerical variations in species exhibiting holocentric chromosomes: A nomenclatural proposal. Caryologia 1996, 49, 301–309. [Google Scholar] [CrossRef]
- Kirschner, J. Karyological differentiation of Luzula sect. Luzula in Europe. Thaiszia 1992, 2, 11–39. Available online: https://www.upjs.sk/public/media/5506/thaiszia-2-11-39-1992-kirschner.pdf (accessed on 15 September 2022).
- Kirschner, J. Taxonomic survey of Luzula sect. Luzula (Juncaceae) in Europe. Folia Geobot. Phytotaxon. 1993, 28, 141–182. [Google Scholar] [CrossRef]
- Bačič, T.; Dolenc Koce, J.; Frajman, B. Diversification and distribution patterns of Luzula sect. Luzula (Juncaceae) in the Eastern Alps: A cytogenetic approach combined with extensive herbarium revisions. Alp. Bot. 2019, 129, 149–161. [Google Scholar] [CrossRef]
- Bačič, T.; Dolenc Koce, J.; Jogan, N. Luzula sect. Luzula (Juncaceae) in the South-Eastern Alps: Morphology, determination and geographic distribution. Bot. Helv. 2007, 117, 75–88. [Google Scholar] [CrossRef]
- Kirschner, J. Luzula . In Juncaceae 1: Rostkovia to Luzula, Species Plantarum: Flora of the World Part 6; Kirschner, J., Ed.; National Library of Australia: Canberra, Australia, 2002; pp. 18–188. [Google Scholar]
- Bačič, T.; Frajman, B.; Dolenc Koce, J. Diversification of Luzula sect. Luzula (Juncaceae) on the Balkan Peninsula—A Cytogenetic Approach. Folia Geobot. 2016, 51, 51–163. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Melters, D.P.; Paliulis, L.V.; Korf, I.F.; Chan, S.W. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012, 20, 579–593. [Google Scholar] [CrossRef]
- Tremetsberger, K.; König, C.; Samuel, R.; Pinsker, W.; Stuessy, T.F. Infraspecific genetic variation in Biscutella laevigata (Brassicaceae): New focus on Irene Manton’s hypothesis. Plant Syst. Evol. 2002, 233, 163–181. [Google Scholar] [CrossRef]
- Parisod, C.; Besnard, G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae). Mol. Ecol. 2007, 16, 2755–2767. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, P.; Lumaret, R.; Bédécarrats, A. Genetic variation and gene flow in Alpine diploid and tetraploid populations of Lotus (L. alpinus (D.C.) Schleicher/L. corniculatus L.). I. Insights from morphological and allozyme markers. Heredity 1998, 80, 683–693. [Google Scholar] [CrossRef]
- Winkler, M.; Escobar Garcia, P.; Gattringer, A.; Sonnleitner, M.; Huelber, K.; Schönswetter, P.; Schneeweiss, G.M. A novel method to infer the origin of polyploids from AFLP data reveals that the Alpine polyploid complex of Senecio carniolicus (Asteraceae) evolved mainly via autopolyploidy. Mol. Ecol. Ressour. 2017, 17, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Zedek, F.; Bureš, P. Holocentric chromosomes: From tolerance to fragmentation to colonization of the land. Ann. Bot. 2018, 121, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Schönswetter, P.; Stehlik, I.; Holderegger, R.; Tribsch, A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol. Ecol. 2005, 14, 3547–3555. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, M.; Hülber, K.; Flatscher, R.; Escobar García, P.; Winkler, M.; Suda, J.; Schönswetter, P.; Schneeweiss, G.M. Ecological differentiation of diploid and polyploid cytotypes of Senecio carniolicus sensu lato (Asteraceae) is stronger in areas of sympatry. Ann. Bot. 2016, 117, 269–276. [Google Scholar] [CrossRef]
- Kolář, F.; Čertner, M.; Suda, J.; Schönswetter, P.; Husband, B.C. Mixed-ploidy species: Progress and opportunities in polyploid research. Trends Plant Sci. 2017, 22, 1041–1055. [Google Scholar] [CrossRef]
- Fischer, M.A.; Oswald, K.; Adler, W. Exkursionsflora für Österreich, Liechtenstein und Südtirol, 3. Auflage; Biologiezentrum der Oberösterreichischen Landesmuseen: Linz, Austria, 2008; pp. 1096–1099. [Google Scholar]
- Suda, J.; Travnicek, P. Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—New prospects for plant research. Cytom. Part A 2006, 69, 273–280. [Google Scholar] [CrossRef]
- Schönswetter, P.; Suda, J.; Popp, M.; Weiss-Schneeweiss, H.; Brochmann, C. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Mol. Phylogenetics Evol. 2007, 42, 92–103. [Google Scholar] [CrossRef]
- R, version 4.2.2; a Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: http://www.R-project.org/ (accessed on 15 September 2022).
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Schönswetter, P.; Solstad, H.; García, P.E.; Elven, R. A combined molecular and morphological approach to the taxonomically intricate European mountain plant Papaver alpinum s.l. (Papaveraceae)—Taxa or informal phylogeographical groups? Taxon 2009, 58, 1326–1343. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Frajman, B.; Graniszewska, M.; Schönswetter, P. Evolutionary patterns and morphological diversification within the European members of the Euphorbia illirica (E. villosa) group: One or several species? Preslia 2016, 88, 369–390. [Google Scholar]
- Arrigo, N.; Tuszynski, J.W.; Ehrich, D.; Gerdes, T.; Alvarez, N. Evaluating the impact of scoring parameters on the structure of intra-specific genetic variation using RawGeno, an R package for automating AFLP scoring. BMC Bioinform. 2009, 10, 33. [Google Scholar] [CrossRef]
- R, version 3.3.2; a Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://cran.r-project.org (accessed on 12 March 2018).
- Bonin, A.; Bellemain, E.; Bronken Eidesen, P.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261–3273. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Oksanen, F.J.; Blanchet, G.F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.5-4. 2019. Available online: https://github.com/vegandevs/vegan (accessed on 15 September 2022).
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-Means Clustering Algorithm. J. R. Stat. Soc. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Arrigo, N.; Felber, F.; Parisod, C.; Buerki, S.; Alvarez, N.; David, J.; Guadagnuolo, R. Origin and expansion of the allotetraploid Aegilops geniculata, a wild relative of wheat. New Phytol. 2010, 187, 1170–1180. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pungaršek, Š.; Dolenc Koce, J.; Bačič, M.; Barfuss, M.H.J.; Schönswetter, P.; Frajman, B. Disentangling Relationships among the Alpine Species of Luzula Sect. Luzula (Juncaceae) in the Eastern Alps. Plants 2023, 12, 973. https://doi.org/10.3390/plants12040973
Pungaršek Š, Dolenc Koce J, Bačič M, Barfuss MHJ, Schönswetter P, Frajman B. Disentangling Relationships among the Alpine Species of Luzula Sect. Luzula (Juncaceae) in the Eastern Alps. Plants. 2023; 12(4):973. https://doi.org/10.3390/plants12040973
Chicago/Turabian StylePungaršek, Špela, Jasna Dolenc Koce, Martina Bačič, Michael H. J. Barfuss, Peter Schönswetter, and Božo Frajman. 2023. "Disentangling Relationships among the Alpine Species of Luzula Sect. Luzula (Juncaceae) in the Eastern Alps" Plants 12, no. 4: 973. https://doi.org/10.3390/plants12040973
APA StylePungaršek, Š., Dolenc Koce, J., Bačič, M., Barfuss, M. H. J., Schönswetter, P., & Frajman, B. (2023). Disentangling Relationships among the Alpine Species of Luzula Sect. Luzula (Juncaceae) in the Eastern Alps. Plants, 12(4), 973. https://doi.org/10.3390/plants12040973





