On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana
Abstract
1. Introduction
2. Structure of TBP
3. Structure and Variation of the TATA Box
4. Assembly of the Preinitiation Complex
5. Comparative Structural Analysis of Core Promoters of A. thaliana
6. Mutations in TATA Boxes of A. thaliana
7. Structural and Functional Roles of A. thaliana TBP1 and TBP2
8. Participation of TBP in Other Cellular Processes
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levine, M.; Tjian, R. Transcription regulation and animal diversity. Nature 2003, 424, 147–151. [Google Scholar] [CrossRef]
- Levine, M. Paused RNA polymerase II as a developmental checkpoint. Cell 2011, 145, 502–511. [Google Scholar] [CrossRef]
- Werner, F.; Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011, 9, 85–98. [Google Scholar] [CrossRef]
- Zhou, M.; Law, J.A. RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II’s rules. Curr. Opin. Plant Biol. 2015, 27, 154–164. [Google Scholar] [CrossRef]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef]
- Aso, T.; Conaway, J.W.; Conaway, R.C. Role of core promoter structure in assembly of the RNA polymerase II preinitiation complex. A common pathway for formation of preinitiation intermediates at many TATA and TATA-less promotersю. J. Biol. Chem. 1994, 269, 26575–26583. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Burley, S.K. RNA polymerase II transcription initiation: A structural view. Proc. Natl. Acad. Sci. USA 1997, 94, 15–22. [Google Scholar] [CrossRef]
- Berk, A.J. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 1999, 11, 330–335. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229. [Google Scholar] [CrossRef]
- Sandelin, A.; Carninci, P.; Lenhard, B.; Ponjavic, J.; Hayashizaki, Y.; Hume, D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet. 2007, 8, 424–436. [Google Scholar] [CrossRef]
- Roy, A.; Singer, D.S. Core promoters in transcription: Old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. [Google Scholar] [CrossRef]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Dreos, R.; Sloutskin, A.; Malachi, N.; Ideses, D.; Bucher, P. Computational identification and experimental characterization of preferred downstream positions in human core promoters. PLoS Comput. Biol. 2021, 17, e1009256. [Google Scholar] [CrossRef]
- Burke, T.W.; Kadonaga, J.T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997, 11, 3020–3031. [Google Scholar] [CrossRef]
- Sloutskin, A.; Shir-Shapira, H.; Freiman, R.N.; Juven-Gershon, T. The Core Promoter Is a Regulatory Hub for Developmental Gene Expression. Front. Cell Dev. Biol. 2021, 9, 666508. [Google Scholar] [CrossRef]
- Louder, R.K.; He, Y.; López-Blanco, J.; Fang, J.; Chacón, P.; Nogales, E. Structure of promoter-bound TFIID and insight into human PIC assembly. Nature 2016, 531, 604–609. [Google Scholar] [CrossRef]
- Gershenzon, N.I.; Ioshikhes, I.P. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 2005, 21, 1295–1300. [Google Scholar] [CrossRef]
- Tokusumi, Y.; Ma, Y.; Song, X.; Jacobson, R.H.; Takada, S. The New Core Promoter Element XCPE1 (X Core Promoter Element 1) Directs Activator-, Mediator-, and TATA-Binding Protein-Dependent but TFIID-Independent RNA Polymerase II Transcription from TATA-Less Promoters. Mol. Cell Biol. 2007, 27, 1844–1858. [Google Scholar] [CrossRef]
- Anish, R.; Hossain, M.B.; Jacobson, R.H.; Takada, S. Characterization of transcription from TATA-less promoters: Identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS ONE 2009, 4, e5103. [Google Scholar] [CrossRef]
- Cox, J.M.; Hayward, M.M.; Sanchez, J.F.; Gegnas, L.D.; van der Zee, S.; Dennis, J.H.; Sigler, P.B.; Schepartz, A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc. Natl. Acad. Sci. USA 1997, 94, 13475–13480. [Google Scholar] [CrossRef]
- Lagrange, T.; Kapanidis, A.N.; Tang, H.; Reinberg, D.; Ebright, R.H. New core promoter element in RNA polymerase II-dependent transcrition: Sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998, 12, 34–44. [Google Scholar] [CrossRef]
- Deng, W.; Roberts, S.G. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423. [Google Scholar] [CrossRef]
- Buratowski, S.; Hahn, S.; Guarente, L.; Sharp, P.A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 1989, 56, 549–561. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Chen, H.; Halay, E.D.; Usheva, A.A.; Hisatake, K.; Lee, D.K.; Roeder, R.G.; Burley, S.K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 1995, 377, 119–128. [Google Scholar] [CrossRef]
- Buratowski, S.; Zhou, H. Functional domains of transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 1993, 90, 5633–5637. [Google Scholar] [CrossRef]
- Chen, H.T.; Warfield, L.; Hahn, S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007, 14, 696–703. [Google Scholar] [CrossRef]
- Bushnell, D.A.; Westover, K.D.; Davis, R.E.; Kornberg, R.D. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 2004, 303, 983–988. [Google Scholar] [CrossRef]
- Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Wiley interdisciplinary reviews. Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef]
- Burley, S.K.; Roeder, R.G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 1996, 65, 769–799. [Google Scholar] [CrossRef]
- Lee, S.; Hahn, S. Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature 1995, 376, 609–612. [Google Scholar] [CrossRef]
- Bernués, J.; Carrera, P.; Azorin, F. TBP binds the transcriptionally inactive TA5 sequence but the resulting complex is not efficiently recognised by TFIIB and TFIIA. Nucleic Acids Res. 1996, 24, 2950–2958. [Google Scholar] [CrossRef][Green Version]
- Pugh, B.F. Control of gene expression through regulation of the TATA-binding protein. Gene 2000, 255, 1–14. [Google Scholar] [CrossRef]
- Patikoglou, G.A.; Joseph, L.; Kim, J.L.; Sun, L.; Yang, S.-H.; Kodadek, T.; Burley, S.K. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999, 13, 3217–3230. [Google Scholar] [CrossRef]
- Nogales, E.; Louder, R.K.; He, Y. Structural Insights into the Eukaryotic Transcription Initiation Machinery. Annu. Rev. Biophys. 2017, 46, 59–83. [Google Scholar] [CrossRef]
- Moore, P.A.; Ozer, J.; Salunek, M.; Jan, G.; Zerby, D.; Campbell, S.; Lieberman, P.M. A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol. Cell Biol. 1999, 19, 7610–7620. [Google Scholar] [CrossRef]
- Zehavi, Y.; Kedmi, A.; Ideses, D.; Juven-Gershon, T. TRF2: TRansForming the view of general transcription factors. Transcription 2015, 6, 1–6. [Google Scholar] [CrossRef]
- Müller, F.; Zaucker, A.; Tora, L. Developmental regulation of transcription initiation: More than just changing the actors. Curr. Opin. Genet. Dev. 2010, 20, 533–540. [Google Scholar] [CrossRef]
- Hoffman, A.; Sinn, E.; Yamamoto, T.; Wang, J.; Roy, A.; Horikoshi, M.; Roeder, R.G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 1990, 346, 387–390. [Google Scholar] [CrossRef]
- Meisterernst, M.; Roeder, R.G. Family of proteins that interact with TFIID and regulate promoter activity. Cell 1991, 67, 557–567. [Google Scholar] [CrossRef]
- Horikoshi, M.; Yamamoto, T.; Ohkuma, Y.; Weil, P.A.; Roeder, R.G. Analysis of structure-function relationships of yeast TATA box binding factor TFIID. Cell 1990, 61, 1171–1178. [Google Scholar] [CrossRef]
- Horikoshi, M.; Bertuccioli, C.; Takada, R.; Wang, J.; Yamamoto, T.; Roeder, R.G. Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc. Natl. Acad. Sci. USA 1992, 89, 1060–1064. [Google Scholar] [CrossRef]
- Nikolov, D.B.; Hu, S.H.; Lin, J.; Gasch, A.; Hoffmann, A.; Horikoshi, M.; Chua, N.H.; Roeder, R.G.; Burley, S.K. Crystal structure of TFIID TATA-box binding protein. Nature 1992, 360, 40–46. [Google Scholar] [CrossRef]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box comple. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef]
- Kim, J.L.; Nikolov, D.B.; Burley, S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 1993, 365, 520–527. [Google Scholar] [CrossRef]
- Blair, R.H.; Goodrich, J.A.; Kugel, J.F. Single-molecule fluorescence resonance energy transfer shows uniformity in TATA binding protein-induced DNA bending and heterogeneity in bending kinetics. Biochemistry 2012, 51, 7444–7455. [Google Scholar] [CrossRef]
- Wu, J.; Parkhurst, K.M.; Powell, R.M.; Brenowitz, M.; Parkhurst, L.J. DNA bends in TATA-binding protein-TATA complexes in solution are DNA sequence-dependent. J. Biol. Chem. 2001, 276, 14614–14622. [Google Scholar] [CrossRef]
- Faiger, H.; Ivanchenko, M.; Haran, T.E. Nearest-neighbor non-additivity versus long-range non-additivity in TATA-box structure and its implications for TBP-binding mechanism. Nucl. Acids Res. 2007, 35, 4409–4419. [Google Scholar] [CrossRef][Green Version]
- Goldberg, M.L. Sequence Analysis of Drosophila Histone Genes. Ph.D. Thesis, Stanford University, Stanford, CA, USA, 1979. [Google Scholar]
- Yang, C.; Bolotin, E.; Jiang, T.; Sladek, F.M.; Martinez, E. Prevalence of the Initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 2007, 389, 52–65. [Google Scholar] [CrossRef]
- FitzGerald, P.C.; Sturgill, D.; Shyakhtenko, A.; Oliver, B.; Vinson, C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006, 7, R53. [Google Scholar] [CrossRef]
- Shi, W.; Zhou, W. Frequency distribution of TATA Box and extension sequences on human promoters. BMC Bioinform. 2006, 7, S2. [Google Scholar] [CrossRef]
- Bernard, V.; Brunaud, V.; Lecharn, A. TC-motifs at the TATA-box expected position in plant genes: A novel class of motifs involved in the transcription regulation. BMC Genom. 2010, 11, 166. [Google Scholar] [CrossRef]
- Molina, K.; Grotewold, E. Genome wide analysis of Arabidopsis core promoters. BMC Genom. 2005, 6, 25. [Google Scholar] [CrossRef]
- Civan, P.; Svec, M. Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y Patch promoter elements. Genome. 2009, 52, 294–297. [Google Scholar] [CrossRef]
- Bae, S.-H.; Han, H.W.; Moon, J. Functional Analysis of the Molecular Interactions of TATA Box-Containing Genes and Essential Genes. PLoS ONE 2015, 10, e0120848. [Google Scholar] [CrossRef]
- Basehoar, A.D.; Zanton, S.J.; Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 2004, 116, 699–709. [Google Scholar] [CrossRef]
- Bucher, P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990, 212, 563–578. [Google Scholar] [CrossRef]
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117. [Google Scholar] [CrossRef]
- Zuo, Y.-C.; Li, Q.-Z. Identification of TATA and TATA-less promoters in plant genomes by integrating diversity measure, GC-Skew and DNA geometric flexibility. Genomics 2011, 97, 112–120. [Google Scholar] [CrossRef]
- Kimura, K.; Wakamatsu, A.; Suzuki, Y.; Ota, T.; Nishikawa, T.; Yamashita, R.; Yamamoto, J.; Sekine, M.; Tsuritani, K.; Wakaguri, H.; et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006, 16, 55–65. [Google Scholar] [CrossRef]
- Cooper, S.J.; Trinklein, N.D.; Anton, E.D.; Nguyen, L.; Myers, R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006, 16, 1–10. [Google Scholar] [CrossRef]
- Rhee, H.S.; Pugh, B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012, 483, 295–301. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef]
- Gutiérrez, G.; Millán-Zambrano, G.; Medina, D.A.; Jordán-Pla, A.; Pérez-Ortín, J.E.; Peñate, X.; Chávez, S. Subtracting the sequence bias from partially digested MNase-seq data reveals a general contribution of TFIIS to nucleosome positioning. Epigenetics Chromatin 2017, 10, 58. [Google Scholar] [CrossRef]
- Watanabe, K.; Kokubo, T. SAGA mediates transcription from the TATA-like element independently of Taf1p/TFIID but dependent on core promoter structures in Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0188435. [Google Scholar] [CrossRef]
- de Jonge, W.J.; O’Duibhir, E.; Lijnzaad, P.; van Leenen, D.; Groot Koerkamp, M.J.; Kemmeren, P.; Holstege, F.C. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J. 2017, 36, 274–290. [Google Scholar] [CrossRef]
- Baptista, T.; Grünberg, S.; Minoungou, N.; Koster, M.J.E.; Timmers, H.T.M.; Hahn, S.; Devys, D.; Tora, L. SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell 2017, 68, 130–143.e5. [Google Scholar] [CrossRef]
- Berg, O.G.; von Hippel, P.H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 1988, 200, 709–723. [Google Scholar] [CrossRef]
- Savinkova, L.K.; Drachkova, I.A.; Ponomarenko, M.P.; Lysova, M.V.; Arshinova, T.V.; Kolchanov, N.A. Interaction of recombinant TATA-binding protein with TATA-boxes of mammalian gene promoters. Ekol. Genet. 2007, 5, 44–49. [Google Scholar] [CrossRef][Green Version]
- Faiger, H.; Ivanchenko, M.; Cohen, I.; Haran, T.E. TBP flanking sequences: Asymmetry of binding, long-range effects and consensus sequences. Nucleic Acids Res. 2006, 34, 104–119. [Google Scholar] [CrossRef][Green Version]
- Wolner, B.S.; Gralla, J.D. TATA-flanking sequences influence the rate and stability of TATA-binding protein and TFIIB binding. J. Biol. Chem. 2001, 276, 6260–6266. [Google Scholar] [CrossRef]
- Buratowski, S. The basics of basal transcription by RNA polymerase II. Cell 1994, 77, 1–3. [Google Scholar] [CrossRef]
- Parry, T.J.; Theisen, J.W.; Hsu, J.Y.; Wang, Y.L.; Corcoran, D.L.; Eustice, M.; Ohler, U.; Kadonaga, J.T. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010, 24, 2013–2018. [Google Scholar] [CrossRef]
- Lee, D.H.; Gershenzon, N.; Gupta, M.; Ioshikhes, I.P.; Reinberg, D.; Lewis, B.A. Functional characterization of core promoter elements: The downstream core element is recognized by TAF1. Mol. Cell Biol. 2005, 25, 9674–9686. [Google Scholar] [CrossRef]
- Myer, V.E.; Young, R.A. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 1998, 273, 27757–27760. [Google Scholar] [CrossRef]
- Cisse, I.I.; Izeddin, I.; Causse, S.Z.; Boudarene, L.; Senecal, A.; Muresan, L.; Dugast-Darzacq, C.; Hajj, B.; Dahan, M.; Darzacq, X. Real-time dynamics of RNA polymerase II clustering in live human cells. Live-cell imaging visualizes Pol II clusters and their dynamics in human nuclei. Science 2013, 341, 664–667. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Imaging reveals nuclear condensates for Pol II transcription. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A phase separation model for transcriptional control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef]
- Kato, M.; McKnight, S.L. A solid-state conceptualization of information transfer from gene to message to protein. Annu. Rev. Biochem. 2018, 87, 351–390. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 2018, 175, 1842–1855.e1816. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; John C Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Il’icheva, I.A.; Khodikov, M.V.; Poptsova, M.S.; Nechipurenko, D.Y.; Grokhovsky, S.L. Structural features of DNA that determine RNA polymerase II core promoter. BMC Genom. 2016, 17, 973. [Google Scholar] [CrossRef]
- Yang, M.Q.; Laflamme, K.; Gotea, V.; Joiner, C.H.; Seidel, N.E.; Wong, C.; Petrykowska, H.M.; Lichtenberg, J.; Lee, S.; Welch, L.; et al. Genome-wide detection of a TFIID localization element from an initial human disease mutation. Nucleic Acids Res. 2011, 39, 2175–2187. [Google Scholar] [CrossRef]
- Yamamoto, Y.Y.; Yoshitsugu, T.; Sakurai, T.; Seki, M.; Shinozaki, K.; Obokata, J. Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 2009, 60, 350–362. [Google Scholar] [CrossRef]
- Bajic, V.B.; Tan, S.L.; Christoffels, A.; Schönbach, C.; Lipovich, L.; Yang, L.; Hofmann, O.; Kruger, A.; Hide, W.; Kai, C.; et al. Mice and men: Their promoter properties. PLoS Genet. 2006, 2, e54. [Google Scholar] [CrossRef]
- Baumann, M.; Pontiller, J.; Ernst, W. Structure and basal transcription complex of RNA polymerase II core promoters in the mammalian genome: An overview. Mol. Biotechnol. 2010, 45, 241–247. [Google Scholar] [CrossRef]
- Yamamoto, Y.Y.; Ichida, H.; Matsui, M.; Obokata, J.; Sakura, T.; Satou, M.; Seki, M.; Shinozaki, K.; Abe, T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genom. 2007, 8, 67. [Google Scholar] [CrossRef]
- Yamamoto, Y.Y.; Ichida, H.; Abe, T.; Suzuki, Y.; Sugano, S.; Obokata, J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007, 35, 6219–6226. [Google Scholar] [CrossRef]
- Kikuchi, S.; Satoh, K.; Nagata, T.; Kawagashira, N.; Doi, K.; Kishimoto, N.; Yazaki, J.; Ishikawa, M.; Yamada, H.; Ooka, H.; et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 2003, 301, 376–379. [Google Scholar] [CrossRef]
- Das, S.; Bansal, M. Variation of gene expression in plants is influenced by gene architecture and structural properties of promoters. PLoS ONE 2019, 14, e0212678. [Google Scholar] [CrossRef]
- Tirosh, I.; Weinberger, A.; Carmi, M.; Barkai, N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006, 38, 830–834. [Google Scholar] [CrossRef]
- Landry, C.R.; Lemos, B.; Rifkin, S.A.; Dickinson, W.J.; Hartl, D.L. Genetic properties influencing the evolvability of gene exression. Science 2007, 317, 118–121. [Google Scholar] [CrossRef]
- Kumari, S.; Ware, D. Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots. PLoS ONE 2013, 8, e79011. [Google Scholar] [CrossRef]
- Mukumoto, F.; Hirose, S.; Imaseki, H.; Yamazaki, K.-I. DNA sequence requirement of a TATA element-binding protein from Arabidopsis for transcription in vitro. Plant Mol. Biol. 1993, 23, 995–1003. [Google Scholar] [CrossRef]
- Heard, D.J.; Kiss, T.; Filipowicz, W. Both Arabidopsis TATA binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: Evidence for gene-specific changes in DNA binding specificity of TBP. EMBO J. 1993, 12, 3519–3528. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Itoh, Y.; Takeda, Y.; Yamazaki, K. TATA sequence requirements for the initiation of transcription for an RNA polymerase II in vitro transcription system from Nicotiana tabacum. Plant Mol. Biol. 1998, 38, 1247–1252. [Google Scholar] [CrossRef]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef]
- Jiao, Y.; Ma, L.; Strickland, E.; Deng, X.W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 2005, 17, 3239–3256. [Google Scholar] [CrossRef]
- Kiran, K.; Ansari, S.A.; Srivastava, R.; Lodhi, N.; Chaturvedi, C.P.; Sawant, S.V.; Tuli, T. The TATA-Box Sequence in the Basal Promoter Contributes to Determining Light-Dependent Gene Expression in Plants. Plant Physiol. 2006, 142, 364–376. [Google Scholar] [CrossRef]
- Joshi, C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987, 15, 6643–6653. [Google Scholar] [CrossRef]
- Ranjan, A.; Ansari, S.A.; Srivastava, R.; Mantri, S.; Asif, M.H.; Sawant, S.V.; Tuli, R. A T9G Mutation in the Prototype TATA-Box TCACTATATATAG Determines Nucleosome Formation and Synergy with Upstream Activator Sequences in Plant Promoters. Plant Physiol. 2009, 151, 2174–2186. [Google Scholar] [CrossRef]
- Zhu, Q.; Dabi, T.; Lamb, C. TATA box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 1995, 7, 1681–1689. [Google Scholar]
- Appel, H.M.; Fescemyer, H.; Ehlting, J.; Weston, D.; Rehrig, E.; Joshi, T.; Xu, D.; Bohlmann, J.; Schultz, J. Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front. Plant Sci. 2014, 5, 565. [Google Scholar] [CrossRef]
- Jores, T.; Tonnies, J.; Wrightsman, T.; Buckler, E.S.; Cuperus, J.T.; Fields, S.; Queitsch, C. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat. Plants 2021, 7, 842–855. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and for Plant Growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef]
- Zhang, M.; Lv, Y.; Wang, Y.; Rose, J.K.C.; Shen, F.; Han, Z.; Zhang, X.; Xu, X.; Wu, T.; Han, Z. TATA Box Insertion Provides a Selection Mechanism Underpinning Adaptations to Fe Deficiency. Plant Physiol. 2017, 173, 715–727. [Google Scholar] [CrossRef]
- Troukhan, M.; Tatarinova, T.; Bouck, J.; Flawell, R.; Alexandrov, N. Genome-wide discovery of cis-elements in promoter sequences using gene expression data. OMICS 2009, 13, 139–151. [Google Scholar] [CrossRef]
- Tatarinova, T.V.; Alexandrov, N.N.; Bouck, J.B.; Feldmann, K.A. GC3 biology in corn, rice, sorghum and other grasses. BMC Genom. 2010, 11, 308. [Google Scholar] [CrossRef]
- Wang, H.C.; Hickey, D.A. Rapid divergence of codon usage patterns within the rice genome. BMC Evol. Biol. 2007, 7, S6. [Google Scholar] [CrossRef]
- Liu, Q.; Dou, S.; Ji, Z.; Xue, Q. Synonymous codon usage and gene function are strongly related in Oryza sativa. Biosystems 2005, 80, 123–131. [Google Scholar] [CrossRef]
- Moshonov, S.; Elfakess, R.; Golan-Mashiach, M.; Sinvani, H.; Dikstein, R. Links between core promoter and basic gene features influence gene expression. BMC Genom. 2008, 9, 92. [Google Scholar] [CrossRef]
- Mingam, A.; Toffano-Nioche, C.; Brunaud, V.; Boudet, N.; Kreis, M.; Lecharny, A. DEAD-box RNA helicases in Arabidopsis thaliana: Establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol. J. 2004, 2, 401–415. [Google Scholar] [CrossRef]
- Blake, W.J.; Balazsi, G.; Kohanski, M.A.; Isaacs, F.J.; Murphy, K.F.; Kuang, Y.; Cantor, C.R.; Walt, D.R.; Collins, J.J. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 2006, 24, 853–865. [Google Scholar] [CrossRef]
- Hoopes, B.C.; LeBlanc, J.F.; Hawley, D.K. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 1998, 277, 1015–1031. [Google Scholar] [CrossRef]
- Wobbe, C.R.; Struhl, K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell Biol. 1990, 10, 3859–3867. [Google Scholar] [CrossRef]
- Gasch, A.; Hoffmann, A.; Horikoshi, M.; Roeder, R.G.; Chua, N.H. Arabidopsis thaliana contains two genes for TFIID. Nature 1990, 346, 390–394. [Google Scholar] [CrossRef]
- Antonova, S.V.; Boeren, J.; Timmers, H.T.M.; Snel, B. Epigenetics and transcription regulation during eukaryotic diversification: The saga of TFIID. Genes Dev. 2019, 33, 888–902. [Google Scholar] [CrossRef]
- Vogel, J.M.; Roth, B.; Cigan, M.; Freeling, M. Expression of the two maize TATA binding protein genes and function of the encoded TBP proteins by complementation in yeast. Plant Cell 1993, 5, 1627–1638. [Google Scholar] [CrossRef]
- Holdsworth, M.J.; Grierson, C.; Schuch, W.; Bevan, M. DNA-binding properties of cloned TATA-binding protein from potato tubers. Plant Mol. Biol. 1992, 19, 455–464. [Google Scholar] [CrossRef]
- Kim, J.L.; Burley, S.K. 1.9 Å resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat. Struct. Biol. 1994, 1, 638–653. [Google Scholar] [CrossRef]
- Kao, C.C.; Lieberman, P.M.; Schmidt, M.C.; Zhou, Q.; Pei, R.; Berk, A.J. Cloning of a transcriptionally active human TATA binding factor. Science 1990, 248, 1646–1650. [Google Scholar] [CrossRef]
- Muhich, M.L.; Iida, C.T.; Horikoshi, M.; Roeder, R.G.; Parker, C.S. cDNA clone encoding Drosophila transcription factor TFIID. Proc. Natl. Acad. Sci. USA 1990, 87, 9148–9152. [Google Scholar] [CrossRef]
- Horikoshi, M.; Wang, C.K.; Fujii, H.; Cromlish, J.A.; Weil, P.A.; Roeder, R.G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature 1989, 341, 299–303. [Google Scholar] [CrossRef]
- Baldwin, D.A.; Gurley, W.B. Isolation and characterization of cDNAs encoding transcription factor IIB from Arabidopsis and soybean. Plant J. 1996, 10, 561–568. [Google Scholar] [CrossRef]
- Chou, S.; Struhl, K. Transcriptional activation by TFIIB mutants that are severely impaired in interaction with promoter DNA and acidic activation domains. Mol. Cell Biol. 1997, 17, 6794–6802. [Google Scholar] [CrossRef]
- Tansey, W.P.; Herr, W. Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science 1997, 275, 829–831. [Google Scholar] [CrossRef]
- Pan, S.; Czarnecka-Verner, E.; Gurley, W.B. Role of the TATA Binding Protein–Transcription Factor IIB Interaction in Supporting Basal and Activated Transcription in Plant Cells. Plant Cell 2000, 12, 125–136. [Google Scholar] [CrossRef][Green Version]
- Li, Y.-F.; Dubois, F.; Zhou, D.-X.; Dubois, F.; Zhou, D.-X. Ectopic expression of TATA box-binding protein induces shoot proliferation in Arabidopsis. FEBS Lett. 2001, 489, 187–191. [Google Scholar] [CrossRef]
- Chaturvedi, C.P.; Lodhi, N.; Ansari, S.A.; Tiwari, S.; Srivastava, R.; Sawant, S.V.; Tuli, R. Mutated TATA-box/TATA binding protein complementation system for regulated transgene expression in tobacco. Plant J. 2007, 50, 917–925. [Google Scholar] [CrossRef]
- Strubin, M.; Struhl, K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 1992, 68, 721–730. [Google Scholar] [CrossRef]
- Bertrand, C.; Benhamed, M.; Li, Y.-F.; Ayadi, M.; Lemonnier, G.; Renou, J.-P.; Delarue, M.; Zhou, D.-X. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1 is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 2005, 280, 1465–1473. [Google Scholar] [CrossRef]
- Lago, C.; Clerici, E.; Mizzi, L.; Colombo, L.; Kater, M.M. TBP-associated factors in Arabidopsis. Gene 2004, 34, 231–241. [Google Scholar] [CrossRef]
- Lawit, S.J.; O’Grady, K.; Gurley, W.B.; Czarnecka-Verner, E. Yeast two-hybrid map of Arabidopsis TFIID. Plant Mol. Biol. 2007, 64, 73–87. [Google Scholar] [CrossRef]
- Kostrewa, D.; Zeller, M.E.; Armache, K.-J.; Seizl, M.; Leike, K.; Thomm, M.; Cramer, P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 2009, 462, 323–330. [Google Scholar] [CrossRef]
- Wu, C. Heat shock transcription factors: Structure and regulation. Annu. Rev. Cell Dev. Biol. 1995, 11, 441–469. [Google Scholar] [CrossRef]
- Lis, J.; Wu, C. Protein traffic on the heat shock promoter: Parking, stalling, and trucking along. Cell 1993, 74, 1–4. [Google Scholar] [CrossRef]
- Morimoto, R.I. Cells in stress: Transcriptional activation of heat shock genes. Science 1993, 259, 1409–1410. [Google Scholar] [CrossRef]
- Reindl, A.; Schöffl, F. Interaction between the Arabidopsis thaliana heat shock transcription factor HSF1 and the TATA binding protein TBP. FEBS Lett. 1998, 436, 318–322. [Google Scholar] [CrossRef]
- Giardina, C.; Perez-Riba, M.; Lis, J.T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992, 6, 2190–2200. [Google Scholar] [CrossRef]
- Balajee, A.S.; Bohr, V.A. Genomic heterogeneity of nucleotide excision repair. Gene 2000, 250, 15–30. [Google Scholar] [CrossRef]
- Bakó, L.; Umeda, M.; Tiburcio, A.F.; Schell, J.; Koncz, C. The VirD2 pilot protein of Agrobacterium-transferred DNA interacts with the TATA box-binding protein and a nuclear protein kinase in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 10108–10113. [Google Scholar] [CrossRef]
- Coin, F.; Frit, F.; Viollet, V.; Salles, B.; Egly, J.-M. TATA Binding Protein Discriminates between Different Lesions on DNA, Resulting in a Transcription Decreas. Mol. Cell Biol. 1998, 18, 3907–3914. [Google Scholar] [CrossRef]
- Ingles, C.J.; Shales, M.; Cress, W.D.; Triezenberg, S.J.; Greenblatt, J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 1991, 351, 588–590. [Google Scholar] [CrossRef]
- Lee, B.C.; Kao, C.C.; Bryant, G.O.; Liu, X.; Berk, A.J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell 1991, 67, 365–376. [Google Scholar] [CrossRef]
- Kashanchi, F.; Piras, G.; Radonovich, M.F.; Duvall, J.F.; Fattaey, A.; Chiang, C.M.; Roeder, R.G.; Brady, J.N. Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature 1994, 367, 295–299. [Google Scholar] [CrossRef]
- Farmer, G.; Colgan, J.; Nakatani, Y.; Manley, J.L.; Prives, C. Functional interaction between p53, the TATA-binding protein (TBP), andTBP-associated factors in vivo. Mol. Cell Biol. 1996, 16, 4295–4304. [Google Scholar] [CrossRef]
- Vichi, P.; Coin, F.; Renaud, J.P.; Vermeulen, W.; Hoeijmakers, J.H.; Moras, D.; Egly, J.M. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997, 16, 7444–7456. [Google Scholar] [CrossRef]
- Auble, D.T.; Hansen, K.E.; Mueller, C.G.; Lane, W.S.; Thorner, J.; Hahn, S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994, 8, 1920–1934. [Google Scholar] [CrossRef][Green Version]
- Auble, D.T.; Wang, D.; Post, K.W.; Hahn, S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell Biol. 1997, 17, 4842–4851. [Google Scholar] [CrossRef]
- Heiss, G.; Ploetz, E.; von Voithenberg, L.V.; Viswanathan, R.; Glaser, S.; Schluesche, P.; Madhira, S.; Meisterernst, M.; Auble, D.T.; Lamb, D.C. Conformational changes and catalytic inefficiency associated with Mot1-mediated TBP-DNA dissociation. Nucleic Acids Res. 2019, 47, 2793–2806. [Google Scholar] [CrossRef]
- Viswanathan, R.; True, J.D.; Auble, D.T. Molecular Mechanism of Mot1, a TATA-binding Protein (TBP)-DNA Dissociating Enzyme. J. Biol. Chem. 2016, 291, 15714–15726. [Google Scholar] [CrossRef]
- Geisberg, J.V.; Struhl, K. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol. Cell 2004, 14, 479–489. [Google Scholar] [CrossRef]
- Zanton, S.J.; Pugh, B.F. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. USA 2004, 101, 16843–16848. [Google Scholar] [CrossRef]
- Andrau, J.C.; Van Oevelen, C.J.C.; Van Teeffelen, H.; Weil, P.A.; Holstege, F.C.P.; Timmers, H.T.M. Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 2002, 21, 5173–5183. [Google Scholar] [CrossRef][Green Version]
- Dasgupta, A.; Darst, R.P.; Martin, K.J.; Afshari, C.A.; Auble, D.T. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 2002, 99, 2666–2671. [Google Scholar] [CrossRef]
- Wollmann, P.; Cui, S.; Viswanathan, R.; Berninghausen, O.; Wells, M.N.; Moldt, M.; Witte, G.; Butryn, A.; Wendler, P.; Beckmann, R.; et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 2011, 475, 403–407. [Google Scholar] [CrossRef]
- Liu, D.; Ishima, R.; Tong, K.I.; Bagby, S.; Kokubo, T.; Muhandiram, D.R.; Kay, L.E.; Nakatani, Y.; Ikura, M. Solution structure of a TBP–TAFII230 Complex. Cell 1998, 94, 573–583. [Google Scholar] [CrossRef]
- Prelich, G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2alpha homolog that has both positive and negative roles in transcription in vivo. Mol. Cell Biol. 1997, 17, 2057–2065. [Google Scholar] [CrossRef]
- Koerkamp, M.G.; Heck, A.J.; Holstege, F.C.; Timmers, H.T. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008, 22, 2359–2369. [Google Scholar] [CrossRef]
- Butryn, A.; Schuller, J.M.; Stoehr, G.; Runge-Wollmann, P.; Förster, F.; Auble, D.T.; Hopfner, K.P. Structural basis for recognition and remodeling of the TBP:DNA:NC2 complex by Mot1. Elife 2015, 10, e07432. [Google Scholar] [CrossRef]
- Teves, S.S.; An, L.; Bhargava-Shah, A.; Xie, L.; Darzacq, X.; Tjian, R. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife 2018, 7, e35621. [Google Scholar] [CrossRef]
- Mishal, R.; Luna-Arias, J.P. Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation. Gene 2022, 833, 146581. [Google Scholar] [CrossRef]
- Lopez-Maury, L.; Marguerat, S.; Bahler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]


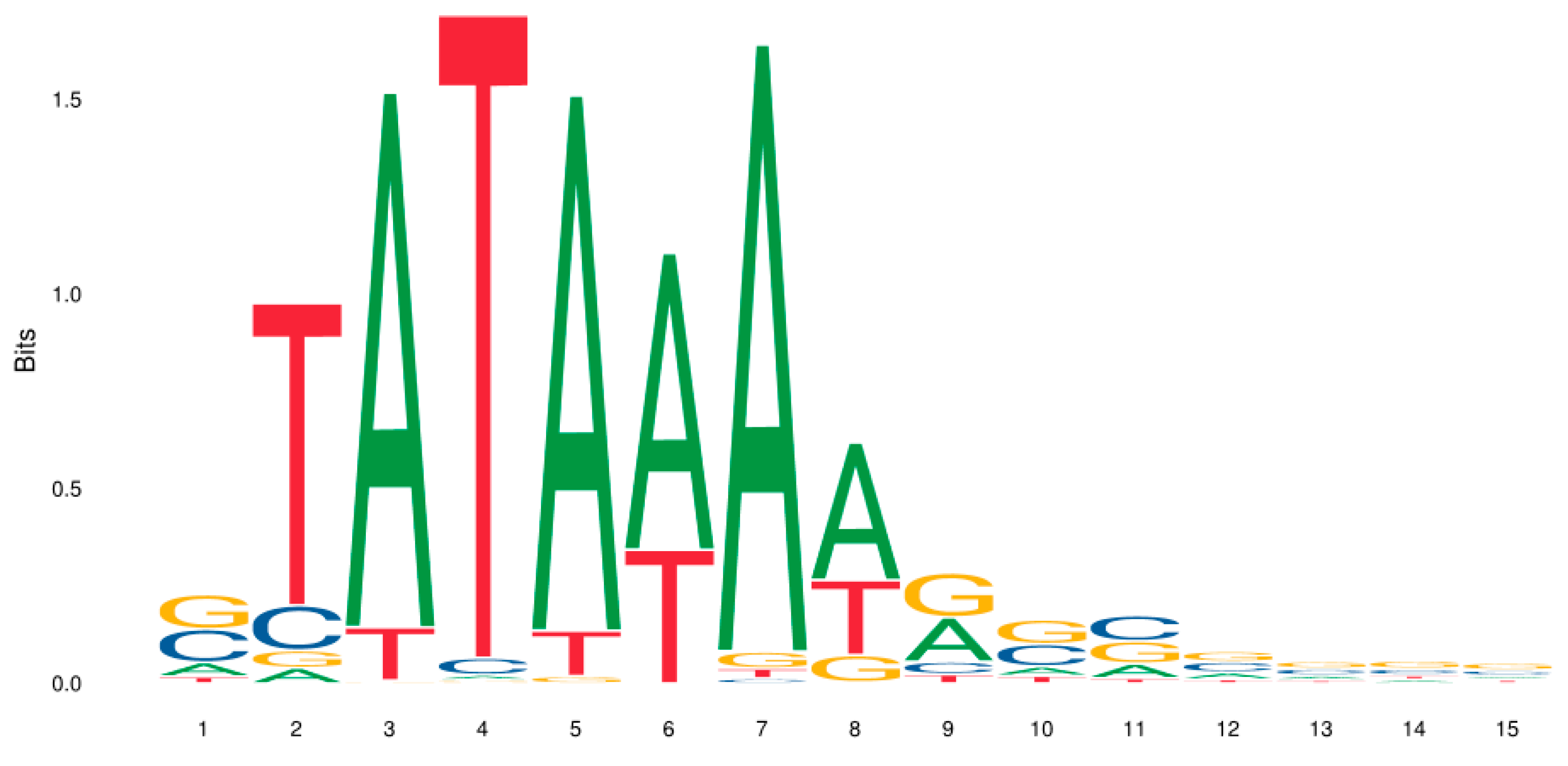


| −3 | −2 | −1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 61 | 16 | 352 | 3 | 354 | 268 | 360 | 222 | 155 | 56 | 83 | 82 | 82 | 68 | 77 |
| C | 145 | 46 | 0 | 10 | 0 | 0 | 3 | 2 | 44 | 135 | 147 | 127 | 118 | 107 | 101 |
| G | 152 | 18 | 2 | 2 | 5 | 0 | 20 | 44 | 157 | 150 | 128 | 128 | 128 | 139 | 140 |
| T | 31 | 309 | 35 | 374 | 30 | 121 | 6 | 121 | 33 | 48 | 31 | 52 | 61 | 75 | 71 |
| G | T | A | T | A | A | A | A | G | G | C | G | C | G | G | |
| C | T | T | T | T | A | C | G | C | C | C | C |
| <2 | <1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | >1 | >2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.28 | 0.16 | 0.03 | 0.95 | 0.00 | 1.00 | 0.62 | 0.97 | 0.38 | 0.73 | 0.13 | 0.30 |
| C | 0.27 | 0.63 | 0.01 | 0.00 | 0.04 | 0.00 | 0.00 | 0.00 | 0.01 | 0.08 | 0.42 | 0.42 |
| G | 0.17 | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.10 | 0.28 | 0.16 |
| T | 0.28 | 0.16 | 0.96 | 0.05 | 0.96 | 0.00 | 0.38 | 0.01 | 0.61 | 0.09 | 0.18 | 0.11 |
| c | T | A | T | A | A/T | A | T/A | A |
| TATA Box | Expression, % |
|---|---|
| TATATATA | 100 |
| TAAATATA | 15 |
| AATATATA | >36 |
| TATAAATA | >36 |
| TATATAAA | >36 |
| TTTATATA | >36 |
| TATTTATA | >36 |
| TATATTTA | >36 |
| TATATATT | >36 |
| TAGAGATA | 0 |
| GAGAGAGA | 0 |
| TATA Box | Expression, % |
|---|---|
| TATAAATA | 100 |
| TACGAATA | 5 |
| TATACGTA | 5 |
| CGTAAATA | 7 |
| TATAAACG | 27 |
| # | TATA Box | Tobacco, % | Drosophila, % | HeLa, % |
|---|---|---|---|---|
| 1 | TATATATA | 100 | 100 | 100 |
| 2 | AATATATA | <36 | ~20 | <60 |
| 3 | CATATATA | <36 | ~17 | <50 |
| 4 | GATATATA | ~20 | ~10 | ~40 |
| 5 | TTTATATA | ~40 | ~15 | >40 |
| 6 | TCTATATA | >30 | ~10 | ~30 |
| 7 | TGTATATA | <20 | <5 | ~30 |
| 8 | TAAATATA | ~7 | 0 | ~17 |
| 9 | TACATATA | >10 | <5 | ~30 |
| 10 | TAGATATA | ~7 | 0 | >30 |
| 11 | TATTTATA | >40 | ~10 | <60 |
| 12 | TATCTATA | <5 | 0 | >20 |
| 13 | TATGTATA | <5 | 0 | ~30 |
| 14 | TATAAATA | 112 | <70 | ~90 |
| 15 | TATACATA | ~5 | <7 | <40 |
| 16 | TATAGATA | <5 | 0 | <40 |
| 17 | TATATTTA | >7 | <7 | ~50 |
| 18 | TATATCTA | >7 | ~10 | ~50 |
| 19 | TATATGTA | >7 | <7 | ~40 |
| 20 | TATATAAA | ~70 | ~110 | ~90 |
| 21 | TATATACA | >7 | <7 | ~50 |
| 22 | TATATAGA | ~30 | ~25 | ~57 |
| 23 | TAGAGATA | 0 | <10 | 0 |
| 24 | GAGAGAGA | 0 | 0 | 0 |
| Promoter Mutation | Dark | Light |
|---|---|---|
| Pmec | 1.04 ± 0.20 | 1.00 |
| T7→C | 0.79 ± 0.16 | 0.01 ± 0.00 |
| T7→G | 2.04 ± 0.40 | 0.02 ± 0.00 |
| A8→C | 0.42 ± 0.08 | 0.01 ± 0.00 |
| A8→G | 1.68 ± 0.32 | 0.02 ± 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savinkova, L.K.; Sharypova, E.B.; Kolchanov, N.A. On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana. Plants 2023, 12, 1000. https://doi.org/10.3390/plants12051000
Savinkova LK, Sharypova EB, Kolchanov NA. On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana. Plants. 2023; 12(5):1000. https://doi.org/10.3390/plants12051000
Chicago/Turabian StyleSavinkova, L. K., E. B. Sharypova, and N. A. Kolchanov. 2023. "On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana" Plants 12, no. 5: 1000. https://doi.org/10.3390/plants12051000
APA StyleSavinkova, L. K., Sharypova, E. B., & Kolchanov, N. A. (2023). On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis thaliana. Plants, 12(5), 1000. https://doi.org/10.3390/plants12051000






